Professor Charlie Carter in the Department of Biochemistry and Biophysics at the University of North Carolina at Chapel Hill looks to the scientific method to understand the origin of genetic coding – and more
Professor Charles Carter trained as a macromolecular X-ray crystallographer with Joseph Kraut at UCSD and with Aaron Klug at the Medical Research Council Laboratory of Molecular Biology in Cambridge, UK.
As the image suggests, the dynamics of the scientific method fascinate Professor Carter. His research has contributed broadly to structural biology, experimental design, mechanistic enzymology – energetic coupling within proteins, bioinformatics, and evolutionary biology – the origin of genetic coding.
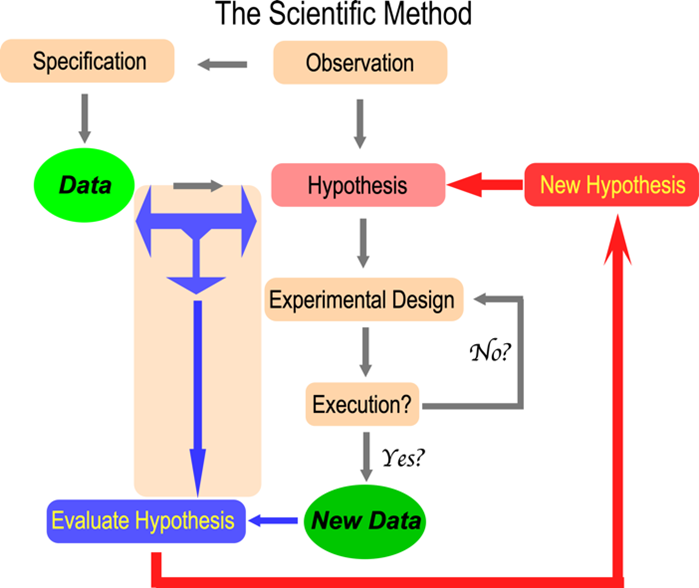
Experimental Design in Research
Most processes of experimental interest depend on multiple factors. Often different factors interact. These interactions are hard to sort out. My father introduced me to factorial experimental design when screening for crystal growth. He taught me to minimize the aliasing of fractional factorial designs. Our paper (1) inspired all subsequent screening for crystal growth. It remains my most cited paper.
Phase permutation (2) established the value of sampling by producing high-accuracy phases using only 1% of the possible combinations. Neal Sloane (Bell Labs) introduced me to response surface experiments, allowing us to explore the neighborhood of optima (3,4). The latter work identified an unexpected aspect of mRNA editing by the cytidine deaminase APOBEC1: a protein co-factor reduced the optimal temperature from 42 C to 37 C, the temperature at which humans live!
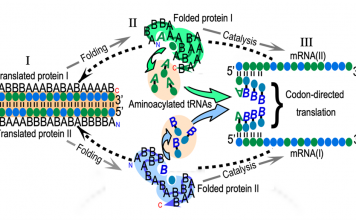
Aminoacyl-tRNA synthetase (aaRS) urzymes and the origin of catalysis and genetic coding
Structural biology studies of tryptophanyl-tRNA synthetase led us to study how the enzymes that translate the genetic code evolved (5,6). We used the protein design program, Rosetta to engineer the most conserved segments of both Class I and II aaRS as soluble catalysts.
The resulting constructs have ~130 residues (7). We call them urzymes. They speed up amino acid activation 109-fold and retain the amino acid activation and tRNA acylation functions necessary to translate a simple code (8).
Protozymes (46-residues) are smaller constructs nested within urzymes. They accelerate amino acid activation 106-fold (9). The catalytic proficiency of the two aaRS Classes increases linearly with sequence length (9). Such parallel improvements are key to synchronizing the different chemical reactions in cells as enzyme sophistication increases.
Several biophysical measurements suggest that urzymes are catalytically active molten globules (10). Thus, the first biological catalysts likely were flexible, partially folded polypeptides that could transiently form tight bonds to chemical reaction transition states.
Bidirectional genetic coding of Class I and II aminoacyl-tRNA synthetases
We confirmed three predictions of the curious hypothesis (11) that ancestral Class I and II aaRS genes were opposite strands of the same gene. (i) The most highly conserved segments (i.e., the urzymes) are precisely those that conform to bidirectional coding.
Their robust catalytic activity is necessary for the hypothesis to be true (5). (ii) Traces of bidirectional coding remain in contemporary aaRS genes. That signal increases as independent reconstructions of the two superfamilies approach the root node (12). (iii) 46-Residue peptides containing the ATP binding sites of Class I and II aaRS, expressed from a designed bidirectional gene have the expected catalytic activities (9).
Moreover, 46-residue peptides have substantial catalytic activity. Peptides from the designed, bidirectional gene achieved the same catalytic proficiency with kcat and KM values 100 times higher than those of the Wild-type WT sequences excerpted from the native structures of Class I TrpRS and Class II HisRS. All four peptides accelerate amino acid activation by ATP by the same amount. Thus, this experiment is a successful direct test of the sense/antisense coding hypothesis of Rodin and Ohno.
Allosteric coupling and free-energy transduction
Domain motion drives amino-acid activation by tryptophanyl-tRNA synthetase. Aromatic side chain repacking in a “master switch” (the D1 switch) mediates the shear forces caused by domain motion. The repacking is rate-limiting. It also enforces multi-state behavior during catalysis. Repacking, in turn, communicates the domain orientation to the active site, activating the active-site Mg2+ ion (13).
Domain motion transiently configures the active site to complement the reaction’s transition state. Combinatorial mutant and modular thermodynamic cycles measure the internal coupling free energies that drive both catalysis (13-15) and specificity (16). Domain movements enhance both catalysis and specificity (16) to the same extent as coupling between individual D1 master switch side chains and the active site (13,14). Dependence of the overall conformational ΔG on PPi release combines with this transient transition state complementarity to use ATP efficiently.
The resulting “escapement mechanism” is a special case of reciprocally-coupled gating, a more general phenomenon, that ensures vectorial transcendence of the second law of thermodynamics (17). It unifies the origins of catalysis, genetics, and bioenergetics (18).
References
- Carter, C.W., Jr. and Carter, C.W. (1979) JBC, 254, 12219-12223.
- Doublié, S., et. al., (1994) Acta Cryst, A50, 164-182.
- Chester, A., et.al., (2004) RNA, 10, 1399 – 1411.
- Yin, Y. and Carter, C.W., Jr. (1994) Acta Cryst., D50, 572-590.
- Carter, C.W., Jr., et. al., (2014) Biology Direct, 9, 11.
- Carter, C.W., Jr. (2014) JBC, 289, 30213–30220.
- Li, L., et. al., (2011) JBC, 286, 10387-10395.
- Li, L., Francklyn, C. and Carter, C.W., Jr. (2013) JBChem, 288, 26856-26863.
- Martinez-Rodriguez, L., et al. (2015) JBC, 290, 19710–19725.
- Tang, G.Q., Hobson, J.J. and Carter, C.W.J. (2023 ) NAR, in review.
- Rodin, S.N. and Ohno, S. (1995) Orig. Life Evol. Biosph., 25, 565-589.
- Chandrasekaran, S.N., et. al., (2013) MBE, 30, 1588-1604.
- Weinreb, V., Li, L. and Carter, C.W., Jr. (2012) Structure, 20, 128-138.
- Weinreb, V., et. al., (2014) JBC 289, 4367-4376.
- Weinreb, V., et. al., (2009) Structure, 17, 1-13.
- Li, L. and Carter, C.W., Jr. (2013) JBC, 288, 34736–34745.
- Carter, C.W., Jr. (2019) Proteins: Structure, Function, and Bioinformatics, 88, 710–717.
- Carter, C.W., Jr. and Wills, P.R. (2021) Biomolecules, 11, 265.