Home 2024
Archives
Structural biology research and the origins of genetic coding
Charles W. Carter, Jr, Department of Biochemistry and Biophysics, University of North Carolina Chapel Hill, reviews the ways that recent research in Structural Biology, Biochemistry, Molecular Biology, and Phylogenetics have opened the origins of genetic coding to experimental study and their important implications.
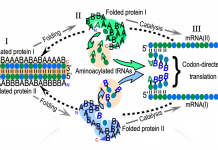
tRNA: The operational RNA code and protein folding
Charles W. Carter, Jr., from the Department of Biochemistry and Biophysics, University of North Carolina at Chapel Hill, relates molecular recognition used in genetic coding to structures of aminoacyl-tRNA synthetases and their cognate tRNAs.
Chicken or egg? Pursuing historical context
Charles W Carter Jr, Department of Biochemistry & Biophysics, University of North Carolina at Chapel Hill, explores prebiotic processes from the historical context enabling the emergence of translation.
AARS urzymes: Experimental biochemistry to map genetic coding
Dr Charlie Carter from the University of North Carolina at Chapel Hill explores how advances in enzymology and phylogenetics enable biochemical measurements that could map the ancestral development of genetic coding.
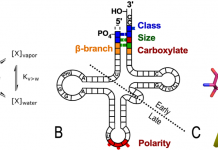
Genetic coding: Roots of genetic readout in nucleic acid structural duality
Charles W. Carter, Jr, from the Department of Biochemistry and Biophysics, University of North Carolina at Chapel Hill explores the roots of genetic readout in the inherent structural duality of DNA and how genetic coding expanded its potential, enabling life to emerge.