Groundbreaking AI models are now deciphering plant DNA, treating genetic sequences like language. This innovation promises to revolutionise genomics and agriculture, offering unprecedented insights into plant biology and accelerating crop improvement for global food security
By leveraging the structural parallels between genomic sequences and natural language, these AI-driven models can decode complex genetic information, offering unprecedented insights into plant biology. This advancement holds promise for accelerating crop improvement, enhancing biodiversity conservation, and bolstering food security in the face of global challenges.
Breaking down genomic barriers with AI
Traditionally, plant genomics has been challenged by the sheer volume and complexity of its datasets. The specificity of traditional machine learning models and the limited availability of annotated data have further complicated progress. However, the advent of large language models (LLMs), which have already revolutionised natural language processing, offers a new approach. A recent study highlights how these models can be adapted to effectively interpret the unique “language” of plant genomes.
Groundbreaking research from Hainan University
A study published in Tropical Plants, details this innovative application of LLMs. Researchers Meiling Zou, Haiwei Chai, and Zhiqiang Xia’s team at Hainan University demonstrated how LLMs, when trained on extensive plant genomic data, can accurately predict gene functions and regulatory elements.
How LLMs understand plant DNA
The study draws parallels between natural language and genomic sequences, training LLMs to understand and predict gene functions, regulatory elements, and expression patterns in plants. Different LLM architectures were explored, including encoder-only models (DNABERT), decoder-only models (DNAGPT), and encoder-decoder models (ENBED). The methodology involved pre-training LLMs on vast datasets of plant genomic sequences and fine-tuning them with specific annotated data. By treating DNA sequences like linguistic sentences, the models identified patterns and relationships within the genetic code.
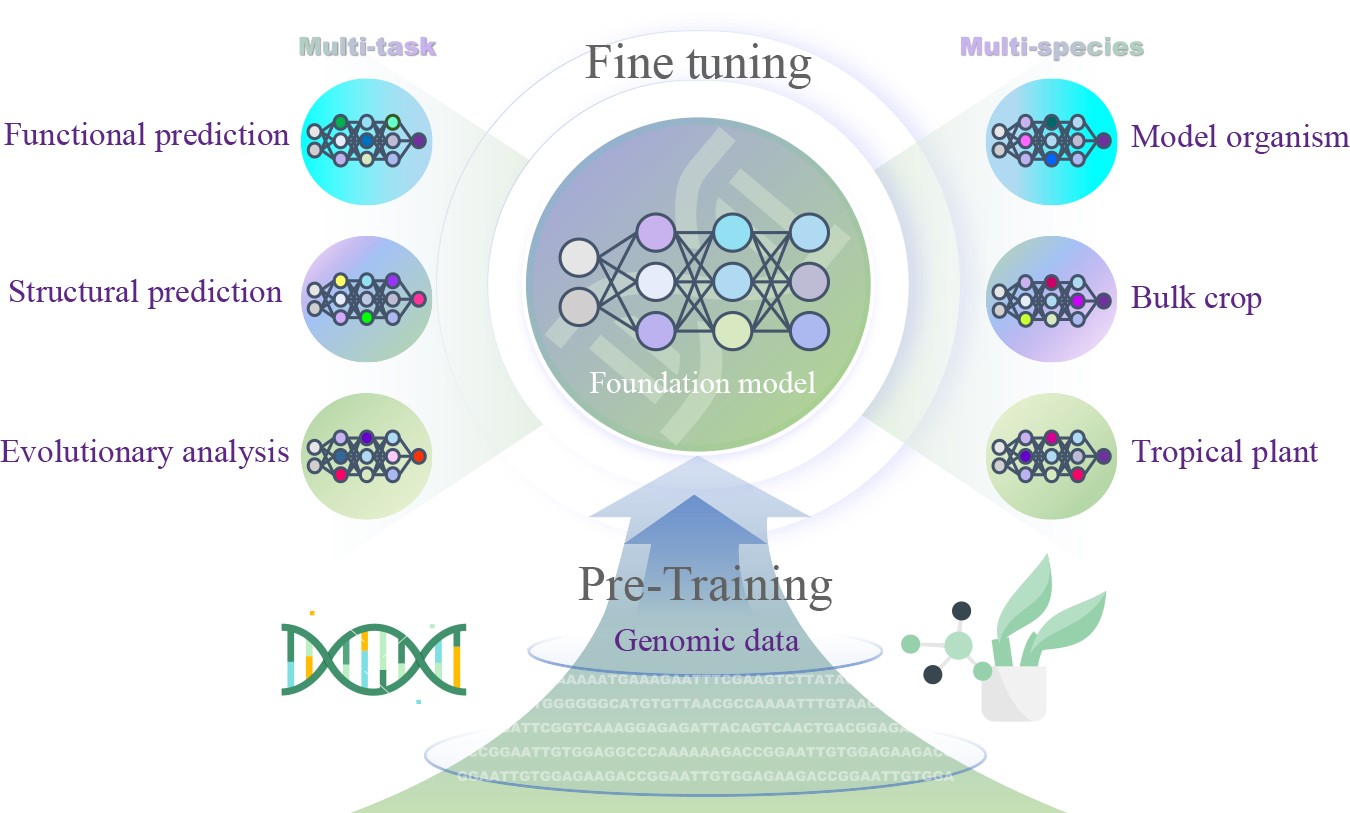
Promising applications and future directions
These models have shown considerable promise in tasks such as promoter prediction, enhancer identification, and gene expression analysis. Plant-specific models like AgroNT and FloraBERT have already demonstrated improved performance in annotating plant genomes and predicting tissue-specific gene expression.
The study also addresses current limitations, noting that most existing LLMs are trained on animal or microbial data, which often lack comprehensive genomic annotations. To overcome this, the authors advocate for the development of plant-focused LLMs trained on diverse plant genomic datasets, including those from underrepresented species like tropical plants. They also emphasise the importance of integrating multi-omics data and developing standardised benchmarks to evaluate model performance.
The future of plant genomics and decoding plant DNA
This research underscores the potential of integrating artificial intelligence, particularly large language models, into plant genomics. By bridging the gap between computational linguistics and genetic analysis, LLMs can revolutionise our understanding of plant biology, paving the way for innovations in agriculture, conservation, and biotechnology. Future research will focus on refining these models, expanding their training datasets, and exploring their applications in real-world agricultural scenarios to fully harness their capabilities.











