In this eBook, Sabine Mai from the University of Manitoba explores how nuclear architecture enables the analysis of genomic instability in cancer
My name is Sabine Mai. I’m a Professor at the University of Manitoba (Winnipeg, Canada). My home department is Physiology and Pathophysiology. I’m also appointed to Biochemistry and Medical Genetics and the Department of Human Anatomy and Cell Science, and I am the Director of the Genomic Center for Cancer Research and Diagnosis, a cutting-edge imaging facility at the University of Manitoba, that I established with funds from the Canada Foundation for Innovation. Additionally, I work as a Senior Investigator at the CancerCare Manitoba Research Institute. I am a Tier 1 Canada Research Chair in Genomic Instability and Nuclear Architecture in Cancer.
Can you explain how nuclear architecture enables the analysis of genomic instability in cancer?
Previously, I have explained the nuclear architecture of the cancer cell using the example of the architecture of a house. When discussing nuclear architecture and genomic instability in cancer, we refer to the nucleus, which contains the genetic information – the chromosomes.
The nucleus has a very specific architecture, which means organization and structure. If you compare that to a house, it becomes very obvious: if you remove parts of the roof from the house, water will collect in some of the rooms when it rains. While this is destructive, the house is still a house. Similarly, genomic instability comes at different levels: low level means that you have some changes/ rearrangements occurring; high level, which is happening in many aggressive tumors, means that you have structural changes that impact the whole function of the cell. Like in the house, when you remove part of the roof or windows, the rain comes in, the wind blows through it and so on.
Genomic instability was described as one of the enabling hallmarks of cancer (Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011 Mar 4;144(5):646-74). Genomic instability is never static. It is always dynamic. So, if there is a genetic change somewhere, unfortunately, it tends to propagate itself and make the genomic instability more severe.
How does the 3D Telomeric Image tool assess the level of genomic instability in cancer cells?
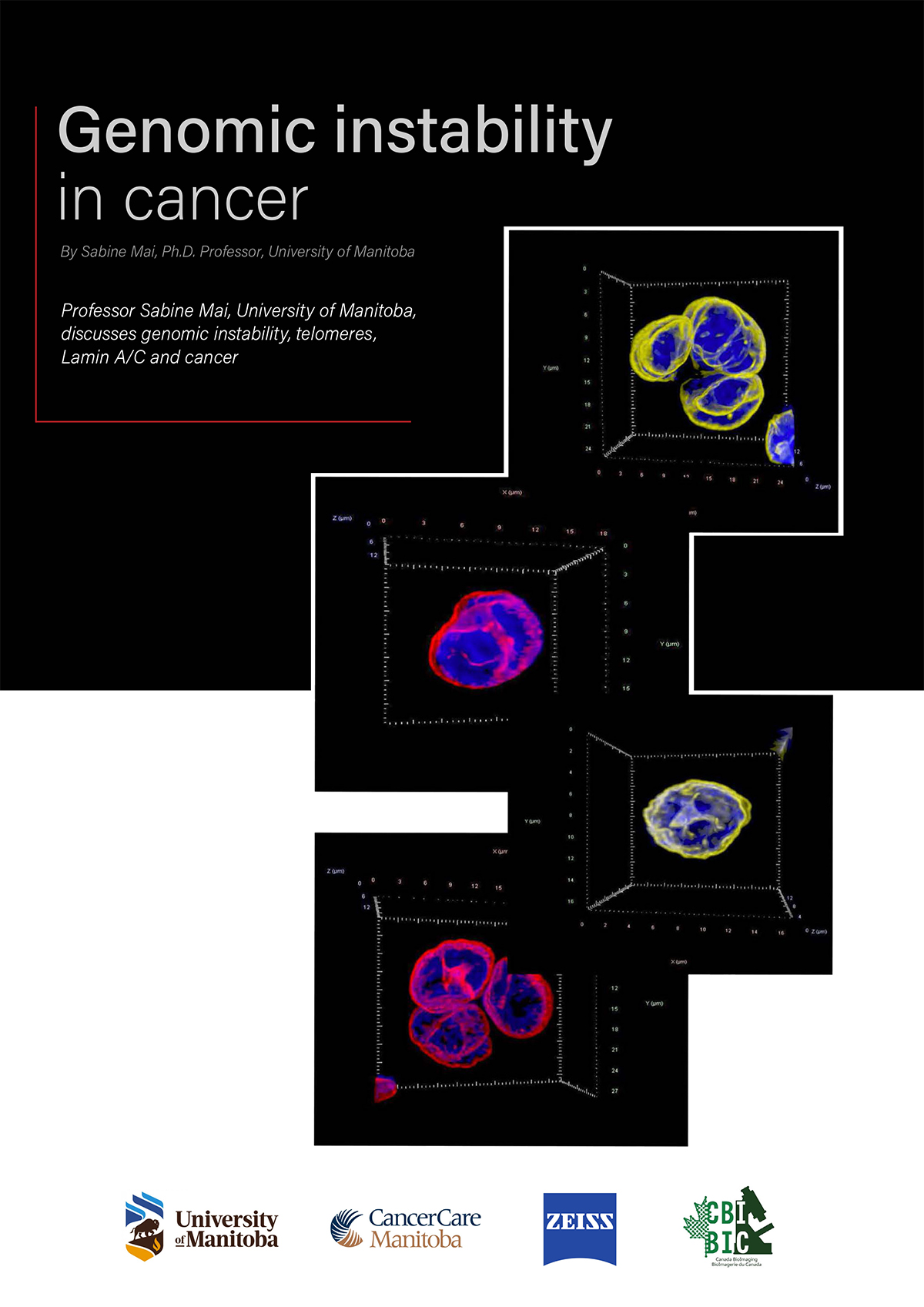
The tool we developed is called TeloView. It is a program that can quantify and measure telomeres (the ends of chromosomes) within the 3D structure of the nucleus. That’s why we created this name (“Telo” from telomeres, “View” because you visualize and quantify). The TeloView software was developed in my academic lab. It is now the property of a company I co-founded called Telo Genomics Corp. It is located in Toronto, Ontario (Canada), at the MaRS Center, the Canadian Innovation and Discovery District.
Importantly, in relation to the importance of TeloView in the clinic, Telo Genomics Corp. uses this program to assess the severity of cancers, the patient’s risk of progression, the patient’s response to treatment, and so on. On the company’s website, readers may view a short video clip summarizing this technology.
Exactly, what does TeloView measure? We just talked about it in your first question, we talked about nuclear architecture. Telomeres are the ends of chromosomes. If we label the ends of each chromosome (telomeres) with a fluorescent dye, then, under a fluorescent microscope, the telomeric ends will emit a fluorescent signal. These signals represent the chromosomal (in)stability present in the nucleus. We have established that the 3D organization of telomeres provides a structural biomarker of the nuclear organization and thus the nuclear architecture.
Why do I say this? In the lab, we have made a comparison between normal cells and tumor cells at various stages, and we have found that there is a very non-random, clear structural organization of telomeres in healthy (control) cells. Whereas in tumor cells, the nuclear architecture is changing.
What are these changes? There are a number of telomeric changes that we measure, and they indicate the level of genomic instability present in the nucleus. The following will summarize the parameters we measure and their meaning.
- Parameter 1.
- We measure the number of telomeres per nucleus. Cancer cells tend to gain and lose chromosomes. If you add a chromosome, that chromosome has its own telomeres, and you have additional signals that you can detect. If you lose a chromosome, you lose signals. The scientific term for gaining or losing of chromosomes is uneuploidy, which means it is not the normal number (not a diploid cell). We thus detect changes in ploidy by measuring telomere numbers.
- Parameter 2.
- We measure the telomeric signal intensity. That is a measure of the length of each individual telomere. Normally, when cells age, the telomeres are shortened. In cancer cells, they may shorten, but the cell will not age and will not die because it has activated maintenance mechanisms. So, measuring the intensity of the telomeres (their length) gives us an overview of the overall genomic instability in the cells.
- Parameter 3.
- We measure the number of telomeric aggregates. The shorter the telomere, the less likely it is capped by protective proteins and the more likely it is to react with other telomeres to cause rearrangements in the cell. In addition, you can have fusions of telomeres. We call the phenomenon ‘telomeric aggregates.’ These aggregates (clusters of telomeres) indicate that short and long telomeres that are in close vicinity can either interact with each other and recombine or they can fuse. When they fuse, then in the next cell division, such fused chromosomes will be stretched from each daughter cell to the other daughter cell until they break. As a result, each daughter cell ends up with a chromosome that has a double-strand break at its end. This double-strand break will react with other chromosomes, and other chromosomes will fuse and break again (so-called breakage-bridge- fusion cycles). You create what I call ongoing dynamic genomic instability.
- Parameter 4.
- We measure the nuclear volume and the number of telomeres per nuclear volume. The nuclear volume often increases in cancer cells. Measuring the telomeric numbers per nuclear volume is important to determine how aberrant the cell has become.
- Parameter 5.
- This parameter is the centre-periphery distribution of the telomeres, which allows us to determine the exact positions of each telomere in the 3D space of the nucleus.
- Parameter 6.
- This is the cell cycle distribution of the telomeres. This parameter indicates whether a cell is resting or actively dividing. The more cells you have in division, the more aggressive your tumor is.
All these parameters taken together allow us to clearly determine on a single cell level how stable or unstable the nucleus is and how much genomic instability is present. Also, because a tumor is not only a single cell, we have the whole tissue or the whole sample that we analyze, and we identify cell-to-cell heterogeneity, which can determine if there are subclones that differ from each other.
How significant is the 3D Telomeric Imaging Tool, and does it have the potential to change the way that we conduct cancer research?
In my opinion, yes. It helps basic and translational research by allowing you to assess the level of genomic instability in multiple cancers. We have examined over 14 different tumor types. And we are able to clearly detect differences between patients, between cells and, of course, differences between normal tissue, normal cells and tumor cells. For cancer research, it’s an important tool, but its use is not exclusive to cancer research.
I see it in precision/personalized medicine. I see it applied in the clinic. The company we founded, Telo Genomics Corp., is doing just that. They have completed clinical trials with the Mayo Clinic and the company will participate at the upcoming American Society of Hematology (ASH) meeting.
Specifically, the company is focused on multiple myeloma, an incurable cancer of the blood. The hope is that when you assess the risk of the patients before myeloma becomes a full-blown disease, one can treat the patient who needs it early and increase the patient’s overall survival.
Can you discuss the function of Lamin/AC in normal and tumor cells and how this relates to genetic instability?
We have previously published some short articles about this topic in OAG and we had a scientific review that was published in Cancers (Dubik N, Mai S. Lamin A/C: Function in Normal and Tumor Cells. Cancers (Basel). 2020 Dec 9;12(12):3688) which both show that Lamin A/C is a very important but not a completely understood player in cancer.
We know that in normal cells, Lamin A/C, provides a scaffold for chromosomes to attach. Imagine a meshwork that has attachment sites for the chromosomes. If that meshwork is changed, it’s either not there or in low abundance or restructured and overexpressed, then this will have an impact on how the chromosomes are organized. And as we know, like in the house that I explained, you have the chairs and the tables in certain places, and when you move them around, you create chaos. The same thing happens in the nuclear architecture of the tumor. It is not yet understood in all detail how Lamin A/C organization affects tumor progression. We have some hints that it may be via creating genomic instability and changing the properties and functions of the tumor cells to make them able to invade tissues where they usually do not belong. But more research needs to be done, and that’s what we are working on.
What does the future hold for the research you’re conducting at the minute?
One cancer that we are focussing on is prostate cancer. We have isolated circulating prostate tumor cells and identified 3D telomeric profiles. These were patients who had not yet received treatment and patients with low-risk, intermediate or high-risk based on clinical classification. Those circulating tumor cells have varying expression levels of Lamin A/C. Some have high, and some have low expression, even within the same patient. There’s a high level of heterogeneity. One aspect that needs to be investigated is whether the level of Lamin A/C found contributes to the patient’s risk of metastasis.
Also in the lab, we have patient-derived prostate cancer cell lines in culture where we have measured the Lamin A/C protein levels and we have found that they can be high, intermediate or low, which is exactly what we found in the circulating tumor cells. A new student who starts in January will investigate how the removal of Lamin A/C would affect the divisions, the proliferation rate and even cell death of prostate cancer cells. We will also look at the chromosome organization, at the nuclear architecture in our attempt to understand if Lamin A/C removal impacts their order and function.
We observed the effects of Lamin A/C on chromosome order in multiple myeloma. One of the students has just finished his Master’s thesis after he established that in multiple myeloma, when you interfere transiently with Lamin A/C protein expression, you disrupt the organization of the chromosomes. He has measured different pairs of chromosomes over time and saw that chromosomes are localized at different positions when Lamin A/C is downregulated. More work needs to be done to understand if Lamin A/C can interfere with cell survival and proliferation rate.
Interested in learning imaging technology? Get in touch
- We offer imaging workshops at the basic, intermediate and advanced levels.
- The basic workshop just concluded (Nov 14-18); please see page 1
- Intermediate and advanced workshops are scheduled for June 2023 and will cover 3D, spectral and super-resolution imaging.
- Please apply if you are interested and contact us at:
- workshop@umanitoba.ca