The expanding availability of high-resolution 3D point clouds from lidar and photogrammetry has fostered interest in digital twin forests
Digital twin forests are datasets in which every tree in a real forest is re-created in a digital setting, including each tree’s specific shape and important biometrics such as tree height, diameter, crown area, and crown volume. With such granular information available for each tree, forest inventories are more accurate, and forestry prescriptions can be tailored for specific demographic or structural outcomes.
An important step in building a digital twin forest is the segmentation of individual trees within the 3D point cloud – that is, determining which 3D points originate from each tree. Diverse algorithms exist to perform tree segmentation, but they share one common challenge: the accidental dismemberment of digital trees prior to segmentation processing.
The challenge: Partial trees confound tree segmentation
Digital tree dismemberment arises because 3D point cloud datasets are usually so large that they must be partitioned into tiles before tree segmentation can be performed using common computing systems. The simplest partitioning approach is to divide the point cloud area into equally sized square or rectangular tiles derived from an overlaid regular XY grid (Fig. 1a).
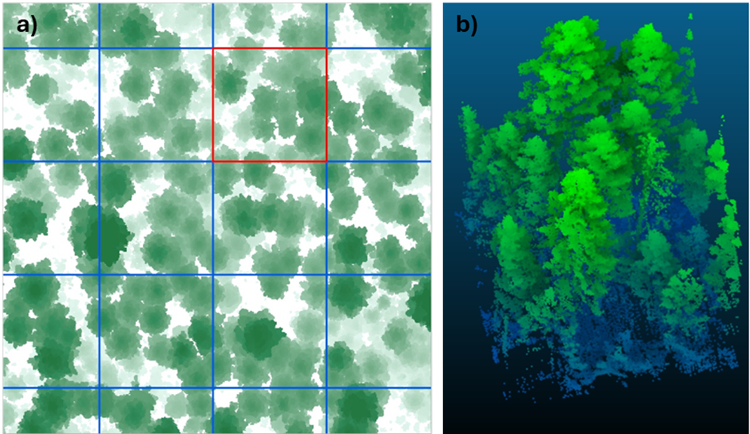
But the straight lines of a regular grid laid across a forest inevitably bisect many trees, meaning that 3D points representing a tree’s stem and crown may get divided among adjacent processing tiles (Fig. 1b).
Understandably, the 3D points from these partial trees scattered around the edges of a tile are processed inconsistently by tree segmentation algorithms, potentially leading to edge trees getting counted more than once or missed entirely, and a high probability that biometrics for any edge tree will be estimated incorrectly due to poor representation of the tree shape. Cleaning up these edge effects in a post-processing step is a considerable challenge, so errors are likely to persist, particularly when manual assessment and correction are not an option.
One solution: From regular grid to irregular grid
An alternative approach is to prevent digital tree dismemberment in the first place by automatically creating an irregular tile grid that does not bisect digital trees. This method takes advantage of a common 2D product of 3D point cloud processing: the canopy height model (CHM). A CHM is a 2D image grid representing the maximum height (i.e., the maximum Z value) of the point cloud across the whole digital forest – effectively an overhead view of tree canopy height (Fig. 2a).
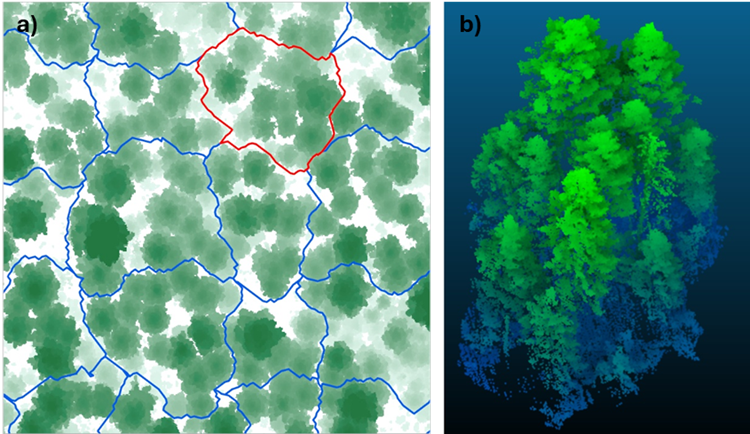
In the irregular grid approach, first a set of grid nodes is distributed regularly across the CHM, but nodes that land on a high value in the CHM are shifted to a nearby low point, to avoid placing nodes within tree canopies. Then, a least-cost path algorithm finds the shortest lines that travel from node to node in both X and Y directions while staying in low areas of the CHM. This means that the lines between grid nodes pass around the tree canopies, instead of cutting across them.
The result is an irregular grid whose lines avoid bisecting trees (Fig. 2b). When the irregular grid is used to partition the 3D point cloud for tree segmentation, the resulting tiles contain points that mainly represent complete trees, ready for evaluation by a segmentation algorithm. With few or no partial trees in the tiles, tile-edge segmentation errors are dramatically reduced, reducing or eliminating post-processing cleanup steps and improving the overall accuracy of the tree segmentation and the final digital twin forest.








