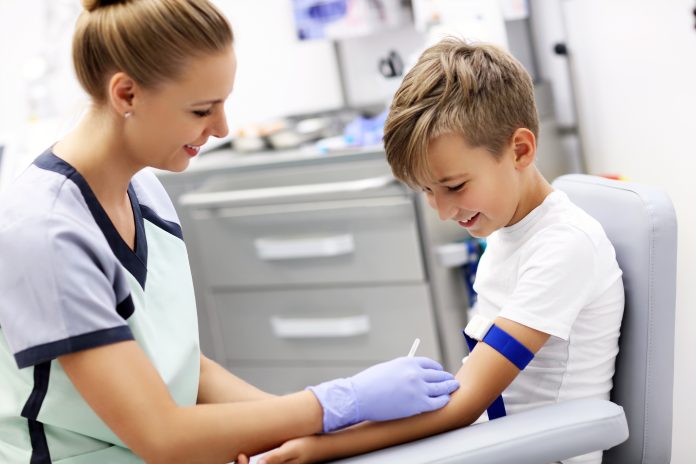
A new rapid blood test, to be presented at the European Society of Human Genetics conference, promises to improve the diagnosis of rare genetic diseases in children significantly
While rare genetic diseases are individually uncommon, there are over 7,000 types caused by mutations in more than 5,000 known genes, collectively affecting around 300 million people worldwide. Currently, about half of all patients with a suspected rare disease remain undiagnosed, and existing testing methods for undiagnosed conditions are typically slow, targeted to a specific disease, and not always sensitive. This can lead to many years of unanswered questions and inconclusive tests.
Dr Daniella Hock, a Senior Postdoctoral Researcher at the University of Melbourne, Australia, and her team have developed a blood-based method of analysing thousands of proteins in a single, untargeted test to improve rare genetic disease diagnosis.
The test rapidly screens thousands of diseases
The proteomic blood test sequences proteins rather than the genes themselves, and the data can help understand how changes in the gene sequence affect its corresponding protein’s function and lead to disease.
The test applies to potentially thousands of different diseases, and it can even detect new ones by providing the evidence needed to confirm that a genetic change is the likely cause of the disease. It is minimally invasive, requiring only one ml of blood from infants, with results available in under three days for patients in acute care.
“When the test is also performed on blood samples from parents, we call it trio analysis. In recessively inherited conditions, this helps considerably in differentiating between carriers, who only have one copy of the defective gene, and the affected individual who carries two copies,” Dr Daniella Hock commented.
New hope for patients with rare genetic diseases
“A recent study carried out in collaboration with the Melbourne School of Population and Global Health revealed that implementing our test in a clinical setting would have a similar cost to that of the current test used to diagnose rare mitochondrial disease, with the advantage that our test can potentially diagnose thousands of other diseases,” added Dr Hock. “Our new test can identify more than 8,000 proteins in peripheral blood mononuclear cells (PBMCs) covering more than 50% of known Mendelian and mitochondrial disease genes, as well as enable us to discover new disease genes.”
The researchers hope their test will become a standard diagnostic procedure for rare genetic diseases in clinical labs.
“The ability to use so little blood from infants and to produce robust results with a rapid turnaround time has been revolutionary to families. Moreover, using familial samples for trio analysis greatly improves the differentiation between carrier and affected individuals with higher confidence, and that has exceeded our initial expectations. We believe that the use of this test in clinical practice will bring considerable benefits to patients, their families and to healthcare systems by reducing the diagnostic time” Dr Hock concluded.
Chair of the conference, Professor Alexandre Reymond, said: “Non-invasive agnostic approaches such as genome sequencing and protein analysis will allow us to reach a diagnosis more rapidly in the future. They will also permit the solving of previously unsolvable cases, thus helping families worldwide.”