Dr Richard E. Goodman of the Food Allergy Research and Resource Program discusses GE crops and the US FDA, EFSA and CODEX international guidelines
Food safety evaluation of genetically engineered (GE) crops and organisms has been debated for decades, before the first GE crops were sold as food in 1996. Some consumers and some scientists who are involved in questions of food safety for a wide variety of plants and animals continue to question whether the safety assessment and regulatory evaluations are sufficient even though there is not a single demonstrated case of food allergy due to any GE organism. Meanwhile food production is not keeping up with increasing demand. Similarly, some people claim there is insufficient evidence of safety and long-term health risks of vaccines to COVID-19 virus. A few people have suffered reactions from the vaccines, but infections can cause death. What evidence is needed for these important developments? Large scale clinical trials were performed with vaccines and now more than 100 million people have been vaccinated. Very few people in the United States have been hospitalized due to severe inflammatory disease cause by the vaccines, but globally more than 3 million have died due to infections.
Some specific GE crops are designed to stop insect pests that cause devastating crop losses in many cases that would normally have been sprayed with chemical pesticides that carry risks of human toxicity and environmental harm. Insect resistant crops and plant-disease resistant crops are rarely possible by conventional crop breeding. The first approved and marketed GE crops included 1) a delayed ripening tomato to improve taste and texture which was dropped after a few years as the benefit was small. 2) Maize (corn) resistant to the European Corn Borer due to the introduction of one gene from a bacterium (Bacillus thuringiensis) that was used safely but inefficiently by organic farmers to stop the same pest for three decades. The Cry 1 Ab is one out of roughly 5,000 proteins encoded by the bacterium. It is insecticidal only to moth caterpillars and is expressed at very low levels (<20 parts per million) in the plant. It has proven very effective for 25 years in controlling the corn borer and can help control Fall Armyworm, a major pest in some countries. The Cry 1 Ab corn is used widely in the US, in Brazil and Argentina, China, and on a limited scale in Spain and Portugal. 3) An herbicide tolerant soybean was produced and sold in 1996 after insertion of a gene encoding the enzyme CP4 EPSPS from another bacterium. The protein takes the place of the natural soybean EPSPS enzyme that encodes production of a molecule essential for aromatic amino acids and other metabolites. The plant enzyme is inactivated by the herbicide glyphosate. This protein is expressed at a modest level of around 400 to 800 parts per million. The soybean is still commonly used in the US, Brazil, Argentina, China and many other countries as a major useful trait that reduces the need for tilling weeds and reduces the need for pre-planting herbicides. The proteins in all three GE products were all evaluated by the safety assessment recommended by the US Food and Drug administration as published in 1992 in the Federal Register following the process in Figure 1. Countries in Europe and India were involved in similar processes in the late 1980’s and early 1990’s. The Food and Agricultural Organization and the World Health Organization of the United Nations sponsored expert panel discussions in 1996, 2000 and 2001 where recommendations for safety evaluations were outlined in Figures 1 and 2. Some steps in Figure 2 are not practical or predictive including a) targeted serum testing, b) animal model testing and c) altered protein digestion protocols.
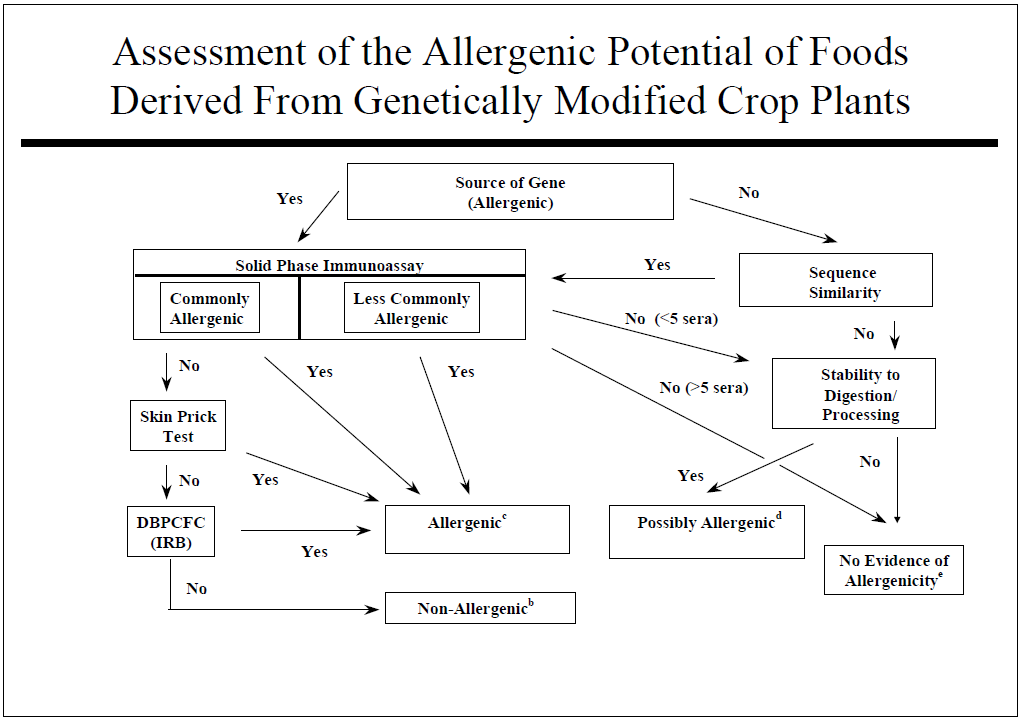
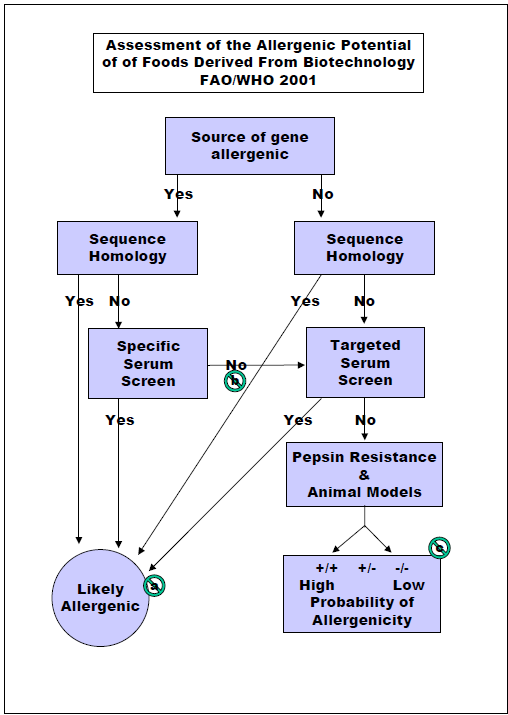
I was hired by Monsanto Company in 1997 with the goal of developing a predictive model for predicting whether a new protein might sensitize consumers and cause them to develop food allergies. The 1996 FAO/WHO document had already outlined a reasonable process and the goal of that was to ensure that a new protein in a GE crop would not cause allergic cross-reactions. When I started testing a mouse model for sensitization by airway inhalation, I was tasked with doing an allergenicity evaluation of insect resistant Bollgard cotton for India. The plant expressed protein was Cry 1 Ac, very similar to Cry 1 Ab in corn. India insisted on using an animal model to predict possible sensitization. We challenged the requirement as no validated animal models were known, but Indian regulators insisted. We used Brown Norway rats feed cotton seed and then tested sensitization using skin prick tests with extracts of cotton seed and control proteins. The test was designed to compare endogenous allergens in cottonseed vs. in GE cotton. No significant differences were identified. India accepted the test results which was conducted in the United States. When Monsanto wanted approval of Bollgard II containing a second insect resistance protein to ensure continued resistance to Pink Bollworm (another moth), Indian regulators demanded the tests be conducted in India. I worked with a commercial toxicology lab in Bangalore, India to conduct the test, but this sensitization was performed by feeding cottonseed meal from Bollgard II for 60 days before testing skin reactivity with extracts of Bollgard II, the parent line and two other commercial non-GE cottonseed lines. The sensitivity to Bollard II extract was similar to the non-transgenic parent cottonseed. Sensitivity to one of the non-GE lines was significantly greater, and the other, significantly lower than Bollgard II. No other country has demanding animal tests and India has dropped that requirement. However, some scientists associated with the European Food Safety Authority (EFSA) are suggesting in 2021 that animal tests should be conducted on any new GE event to predict sensitizing capacity even though a suitable stable model has not been demonstrated to be predictive for human allergy.
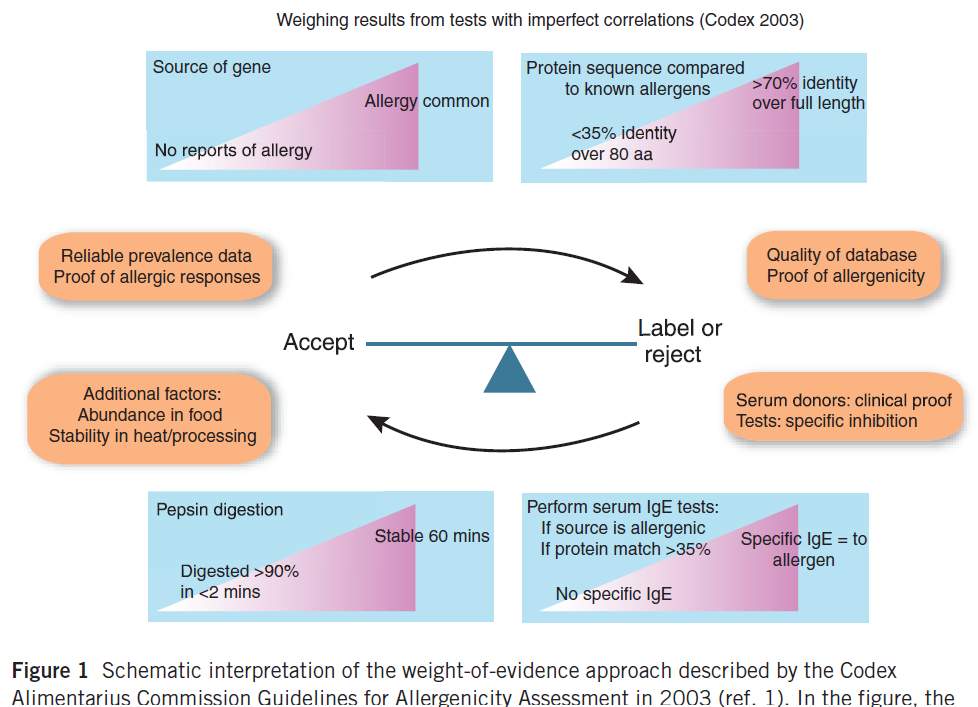
Discussions about improved safety assessments have continued by respected scientists since 2001. The CODEX Commission met September 8 through 12, 2001 after the FAO/WHO 2001 document was published. A number of scientists had written questions and objections to the FAO/WHO 2001 document and those were considered by the CODEX group. The final CODEX document was published in 2003 after CODEX member countries had examined the document closely and voted to accept the final version as a guide for approval of future GE crops, GE microbes and GE animals. Yet now, 20 years later some scientists are still debating methods and pushing to require untested methods into assessments for EFSA and other countries. It is ironic since there is no evidence that an approved GE crop has caused any new allergies or presented any risk of allergic cross-reactivity in 20 years. Since CODEX did not provide a figure, we published an overall evaluation with Figure 3 to help people interpret the process (Goodman et al. 2008).
I joined the University of Nebraska Food Allergy Research and Resource Program in 2004 to develop the www.AllergenOnline.org database specifically for the safety evaluation of GE crops. In 2005 it included 1189 sequences. We update AllergenOnline each year and it now contains 2233 protein sequences from 430 species. The sequences and references are freely available as is the FASTA search using your protein sequence. A sliding 80 amino acid matches was established that includes comparison of shorter identity matches with calculations to adjust to an 80 amino acid length. The rationale for the adjustment is that a sequence of 77 amino acids of the Ara h 2, 2S albumin of peanut would likely cause severe allergic reactions in some people with peanut allergy if it was transformed into a non-peanut crop such as rice. The risk management of food allergy from all sources is based on educating allergic consumers to avoid the foods that cause their allergy. Thus, if a 2S albumin was transferred from peanut into rice, peanut allergic consumers would not realize that they would have to avoid consuming that rice. There are two or three IgE epitopes in the 77 AA segment of Ara h 2 and if their IgE was attached to their mast cells and basophils, they would react after eating that short protein.
Recently we were asked to evaluate predicted proteins of three whole organisms that have not previously been used as food. We used the genome sequences of the organisms to predict protein sequences and those were compared to AllergenOnline.org version 20. The study identified many matches over 35% identity matches over 80 amino acids. We then tested predicted proteins from twenty-three taxonomically diverse species including humans, fish, and common plants. Human protein sequences matched lipid transfer proteins, vicillins, glycinins, tropomyosin and arginine kinase at > 35% identity over 80 amino acids (Abdelmoteleb et al., 2021). Although some homologous proteins from each of the non-human sources are known to be allergens, that does not mean they are all cross-reactive. Testing possible cross-reactivity would require obtaining serum samples from hundreds of allergic consumers, correlating their clinical histories with serum IgE levels and testing potential cross-reactivity in reciprocal fashion. The sequence matches suggest that most consumers would be at risk of allergic cross-reactivity. Yet clinical responses in the allergy literature does not support such broad cross-reactivity (Gray, 2017). Since consumers and regulators are asking questions about the safety of new and novel food sources, we are working to develop methods that are broadly evaluated to assess safety based on sound science. We will be recommending increasing the minimal criteria from >35% identity over 80 to levels based on protein types (2S albumins, vicilins, tropomyosins, etc.) that are predictive based on published likely clinical cross-reactivity. Two challenges are 1) many proteins are highly conserved in evolution with more than 60% identity across broad taxonomic relationships. 2) Humans are exposed to multiple allergenic sources throughout their lives and may be sensitized to two or more protein sources. It is not possible to always predict primary sensitization separately from cross-reactivity. We hope to develop the draft guideline during 2021 for presentation to global regulators since the alternative would be to follow the path of Figure 2 and have a never ended testing cycle for each new or modified food.








