Dana-Farber Cancer Institute has created a blood test that could offer a less invasive way to detect and monitor multiple myeloma
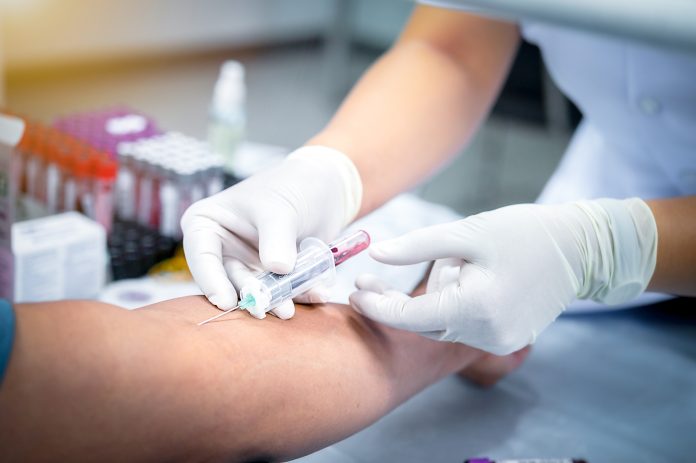
Researchers at Dana-Farber Cancer Institute have developed a new blood test for multiple myeloma that utilises single-cell sequencing to profile circulating tumour cells (CTCs) in the blood. This innovative test offers a non-invasive alternative to traditional bone marrow biopsies, potentially easing the discomfort for patients undergoing regular assessments and for doctors tracking disease progression.
This pioneering development from Dana-Farber Cancer Institute holds great promise for enhancing care and outcomes for individuals grappling with multiple myeloma. By introducing new possibilities for both patients and healthcare providers, this blood test could significantly improve the management of the disease.
“A lot of work has gone into the identification of genomic and transcriptomic features that predict worse outcomes in MM, but we are still lacking the tests to measure them in our patients, said senior author, Dr. Irene M. Ghobrial. “As a clinician, this is the type of next-generation test that I would want to order for my patients.”
The study was published in Nature Cancer.
Transforming multiple myeloma diagnosis
Multiple myeloma is a form of bone marrow cancer, and is traditionally assessed and monitored through bone marrow biopsies. These biopsies are painful, infrequent, and the accompanying technique, Fluorescence in situ hybridisation (FISH), often fails to provide precise results, leading to less effective risk assessment and influencing treatment decisions.
“It would be amazing if we had a blood-based test that can outperform FISH and that works in the majority of patients – we think SWIFT-seq may just be that test,” said Dr. Romanos Sklavenitis-Pistofidis, co-first author.
SWIFT-seq allows doctors to perform risk assessments and genetic monitoring using a blood test, making the process easier and more reliable. The test counts CTCs, provides a detailed genetic profile, and identifies key genetic changes. SWIFT-seq evaluates tumour growth rates and identifies important gene patterns that can predict patient outcomes. This method surpasses the accuracy of bone marrow tests like FISH.
“SWIFT-seq is a powerful option as it can measure the number of CTCs, characterise the genomic alterations of the tumour, estimate the tumour’s proliferative capacity and measure prognostically useful gene signatures in a single test and from a blood sample,” said Ghobrial.
SWIFT-seq produced highly accurate results
The researchers studied 101 patients and healthy donors. They found that SWIFT-seq successfully captured CTCs in 90% of patients with Monoclonal Gammopathy of Undetermined Significance (MGUS) and Smouldering Multiple Myeloma (SMM), three conditions that often precede multiple myeloma.
Notably, the test identified CTCs in 95% of patients with SMM and 94% of patients with newly diagnosed MM, the groups most likely to benefit from improved risk stratification and genomic surveillance. SWIFT-seq’s ability to enumerate CTCs based on the tumour’s molecular barcode, rather than relying on cell surface markers, sets it apart from existing methods like flow cytometry.
“We identified a gene signature that we believe captures the tumour’s circulatory capacity and may partly explain some of the unexplained mysteries of myeloma biology,” said Dr. Elizabeth D. Lightbody, co-first author. “This can have a tremendous impact on how we think about curtailing tumour spread in patients with myeloma and could lead to the development of new drugs for patients.”