Andreas Prokop, Head of the Manchester Fly Facility, explains how fruit fly research generates knowledge important for understanding many human diseases
For 30 years I have been studying the nervous system of the fruit fly Drosophila melanogaster, the tiny insect that hovers over our fruit bowls in summer (Prokop, 2016). You may wonder why anybody would invest professional time or public money in something that seems more of a private hobby than serious research. But I am not alone: the fruit fly has been intensively studied for over 100 years and worldwide over 10,000 scientists are currently estimated to engage in fly research; and their work has great impact: nine (arguably ten) researchers have received a Nobel Prize in ‘Physiology or Medicine’ for their work in Drosophila – the last one as recently as 2017 (Fig. 1). As I will argue here, the biomedical sciences would be very far behind their current status quo without research in fly or other simple organisms, such as the nematode worm C. elegans or baker’s yeast.
Why the fly? A historical perspective
Kick-starting genetics Mere serendipity set in motion the long-lasting interest in the fruit fly: in 1910, studies on evolution by Thomas Hunt Morgan led to the almost accidental finding that genes lie on chromosomes. This started the era of genetics – with Drosophila research leading the field unravelling how genes are organised, become mutated or interact with each other (Allchin, 1997; Brookes, 2001; Kohler, 1994).
Genetics as a tool
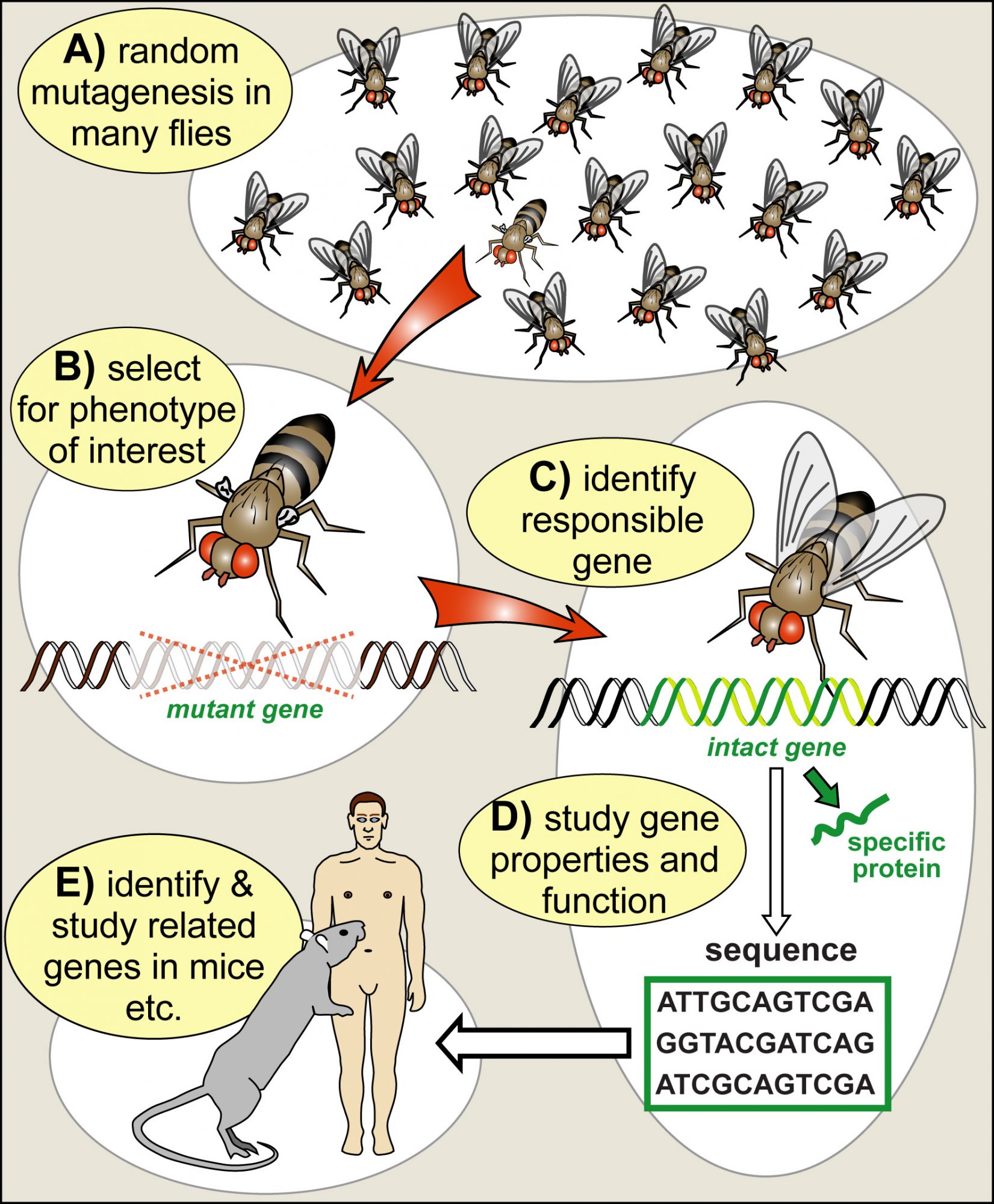
In the middle of the 20th century, researchers started to use Drosophila genetics to address the essential question of how genes work and determine biology. In the same way, as mutations in humans cause inherited diseases that tell us something about the biological relevance of those genes, mutations can be used in Drosophila research as a tool to dissect and understand biological processes. The fly was ideal because genetic manipulation techniques were well established, its generation cycle of only 10 days allowed fast progress, and the ease of keeping big numbers of flies facilitated systematic ‘mutational screens’ to search for new genes that contribute to biological processes (Fig. 2). Drosophila became “a boundary object par excellence, residing in the interstices of two major disciplines, genetics and embryology” (Keller, 1996).
Together with the advent of molecular biology (to decipher and manipulate genes) and advances in biochemistry (to study the protein products of genes), fly research turned into a gold mine for discovery. For example, genes that mediate embryonic development, nervous system function or even the ability to learn were discovered and studied, pioneering fundamental understanding of those processes (Mohr, 2018).

A translational path to humans
Through parallel work in vertebrate animals, in particular the mouse, it became increasingly clear that fundamental concepts discovered in the fly seemed to apply to all animals: genes studied in mammals turned out to be very similar in structure and function to their fly equivalents; in some cases, it was even shown that genes from fly and mouse were interchangeable. The scale of this ‘evolutionary conservation’ became clear when the human and fly genomes were sequenced and compared. Ethan Bier and colleagues reported at the time that 77% of 714 distinct human disease genes matched unique Drosophila sequences (Reiter et al., 2001). The fundamental truth behind this statement was unequivocally documented by a systematic study in yeast using 414 strains with lethal mutations, of which almost half could be ‘cured’ by introducing the equivalent human gene (Kachroo et al., 2015; Leslie, 2015). Therefore, the fundamental processes of biology and the genes involved are ancient; organisms that shared their last common ancestor a billion years ago have maintained many of these fundamental functions to astonishing degrees. This concept of ‘deep homology’ explains the above mentioned Nobel laureates: through their work, they have laid foundations for a fundamental understanding of biological processes which can explain to us what goes wrong in human disease and pave the translational path into the quest for cures.
The importance of Drosophila research is undiminished
The last decades have brought new strategies for research in mice and other vertebrate animals that have now turned also these organisms into true boundary objects. The fairly recent advent of CRISPR technology is seen as the magic silver bullet that has finally closed the experimental gap to research in smaller invertebrate models. However, I would argue that this is a dangerous misconception likely leading to increased research costs, the unnecessary use of animals and a slow-down in scientific advance.
Hugo Bellen was cited to have said: “You get 10 times more biology for a dollar invested in flies than you get in mice” (Levitan, 2015). To illustrate this point, keeping 400 fly stocks requires one stand-alone incubator and £100 a month to pay for food vials and four to six hours of work (Fig. 3); maintaining the same number of mouse strains readily accessible would take at least £12,000 a month and a vast housing facility.
Furthermore, CRISPR technology certainly has enormously accelerated mouse research, but it is also well established in Drosophila and has enhanced the possibilities of fly research to the same degree. Many more arguments can be listed (Prokop, 2015), but I would like to focus here on one last, enormously important aspect: the fact that biology is complex.
Thus, to understand inherited diseases, it is often not sufficient to gain important knowledge of the affected genes and their products; it requires an understanding of the usually complex functional networks in which they operate (Prokop, 2016). An important strategy to unravel complex genetic networks is the simultaneous manipulation of two or more genes in the same individual – a task that is routinely performed in a fly laboratory, but enormously laborious and time-consuming in mice.
Furthermore, experiments, even if based on wellinformed rationale, often fail. In fly, such failure is unfortunate but can be easily absorbed, since time and money invested are usually low, with alternative experiments being set up in a matter of days or weeks rather than months or beyond. Hence, work in fly gives access to flexible experimentation, where try-and-error is a feasible strategy to overcome the challenging enigmas posed by biological complexity.

Conclusions
Understanding biology is the lifeblood for translational research into human disease and, as I have argued here, Drosophila research is a powerful generator of such understanding. Certainly, the fly is NOT a minihuman. For example, it cannot be used to study arthritis or fibrosis, but it can be used to understand fundamental concepts of extracellular matrix regulation underlying those problems. In the context of Alzheimer’s disease, flies are unsuited to study personality loss but can be used to address the still unresolved important question of how this condition triggers nerve cells to die. In any case, the choice of experimental models should always be carefully justified. Consequently, funding panels should, in my opinion, more often question the uses of higher animals where fundamental concepts could be pioneered more efficiently in simpler models – thus spending research money responsibly and speeding up the discovery process.
The author, Andreas Prokop, is Professor for Neurobiology at The University of Manchester. As academic head of the ‘Manchester Fly Facility’ he drives a science communication initiative advocating the wider awareness of fly research. Part of this initiative is the ‘droso4schools’ project aiming to establish Drosophila also as a powerful teaching tool in school biology lessons. The author would like to thank Tom Millard for his helpful comments on the manuscript.
References
Allchin, D. (1997). Thomas Hunt Morgan & the white-eyed mutant. In “Doing Biology (chapter 5)” (J. B. Hagen, D. Allchin, F. Singer, Eds.). Benjamin Cummings — shipseducation.net/db/morgan.htm
Brookes, M. (2001/2002). “Fly: The Unsung Hero of Twentieth-Century Science.” Ecco/Phoenix, – www.amazon.com/Fly-Unsung-Hero-Twentieth-Century-Science/dp/0060936797
Kachroo, A. H., Laurent, J. M., Yellman, C. M., Meyer, A. G., Wilke, C. O., Marcotte, E. M. (2015). Evolution. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science 348, 921-5 – www.ncbi.nlm.nih.gov/pubmed/25999509
Keller, E. F. (1996). Drosophila embryos as transitional objects: the work of Donald Poulson and Christiane Nüsslein-Volhard. Hist Stud Phys Biol Sci 26, 313-46 – www.ncbi.nlm.nih.gov/pubmed/11613313
Kohler, R. E. (1994). “Lords of the fly. Drosophila genetics and the experimental life.” The University of Chicago Press, Chicago, London.
Leslie, M. (2015). Yeast can live with human genes. Blog post in “Science Magazine News” – www.sciencemag.org/news/2015/05/yeast-can-livehuman-genes
Levitan, D. (2015). Paul Knocks flies and NIH funding. Blog post in “FactCheck.org” – www.factcheck.org/2015/02/paul-knocks-flies-andnih-funding
Mohr, S. E. (2018). “First in fly – Drosophila research and biological discovery.” Harvard Univ. Press, Cambridge MA and London UK – www.hup.harvard.edu/catalog.phpisbn=9780674971011
Prokop, A. (2015). Why fly? Blog post in “droso4schools” – droso4schools.wordpress.com/why-fly
Prokop, A. (2016). Fruit flies in biological research. Biological Sciences Review 28, 10-14 – https://t.co/nyz7zxFfl3
Reiter, L. T., Potocki, L., Chien, S., Gribskov, M., Bier, E. (2001). A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res 11, 1114-25 – http://www.ncbi.nlm.nih.gov/pubmed/11381037
Andreas Prokop
Professor of Cellular & Developmental Neurobiology
Manchester Fly Facility
Tel: +44 (0)161 275 1556
Andreas.Prokop@manchester.ac.uk