Decoding pathogens’ genetic material is fast becoming an invaluable tool to support pandemic preparedness and responses to global public health threats, Anona Bamford tells us more
Genomic sequencing and the powerful computational and statistical methods used to analyse the copious amounts of data generated have emerged as crucial tools in predicting and preventing pandemics. By offering valuable insights into the origin, spread and evolution of infectious diseases, scientists can unravel the genetic makeup of pathogens and understand their behaviour, track transmission, and develop targeted interventions.
Whole genome sequencing as a tool towards pandemic preparedness
Whole genome sequencing involves determining an organism’s complete nucleic acid sequence, including viruses and bacteria. In the context of pandemics, it enables scientists to identify the specific genetic signatures associated with a pathogen, providing crucial information about its origin and relatedness to other strains.
Advancements in genomics enabled the global scientific community to rapidly sequence the genome of the SARS- CoV-2 virus, which was instrumental in informing the public health response to COVID-19 worldwide and improving the understanding of how future outbreaks could occur. (1) Analysis of the viral genomic sequences enabled us to decipher transmission patterns, deploy measures to contain the spread of the virus and develop a vaccine within record time. (2) Moreover, genomic sequencing has revolutionised capabilities for pathogen detection and diagnosis.
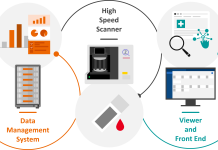
Traditional diagnostic methods have relied in the past on culturing pathogens, which can be time-consuming and often fail to identify novel or fast-evolving strains. In contrast, the availability of the first whole-genome sequences of SARS-CoV-2 facilitated the rapid development of molecular diagnostic techniques, particularly nucleic acid-based diagnostic assays such as real-time reverse transcription-polymerase chain reaction (rRT–PCR).
Molecular testing
Some advantages of molecular rRT-PCR testing include its clinical and analytical sensitivity and specificity and its scalability when transferring such protocols to automated liquid handling workstations to improve workflow optimisation, performance, throughput, and sample-to-result turnaround times. Additionally, samples could be collected remotely by healthcare professionals or self-collected and sent to centralised laboratory testing hubs such as the UK Lighthouse Lab Network – a partnership between Medicines Discovery Catapult (MDC), the Department of Health and Social Care, UK Biocentre, and the University of Glasgow, closely supported by both the NHS and Public Health England. (3) Molecular PCR testing could also indicate some COVID variants ahead of genomic sequencing. (4) This helped expedite our understanding of the prevalence of circulating COVID-19 strains which, in turn, informed the next steps in pandemic preparedness and prediction within communities.
Scientists could reconstruct the phylogenetic tree by comparing viral genomes obtained from different patients, representing the evolutionary relationships among strains. Identification of transmission clusters and tracking the introduction and spread of COVID-19 within a population and across geographical regions was monitored globally by the World Health Organization (WHO). (5)
Sharing (data) is caring
Indeed, as per the WHO recommendation, countries were asked to share genomic data, which further helped develop a coordinated global response against the virus and predict local and global transmission routes. Such data collaborations have been in place for influenza since 2006, when the Global Initiative on Sharing All Influenza Data (GISAID) was set up to facilitate the rapid sharing of influenza genomic data between researchers worldwide. (6)
In March 2020, the COVID-19 Genomics UK (COG-UK) consortium was created to rapidly develop a national capability for SARS-CoV-2 sequencing to inform public health responses. (7) In less than a month, COG-UK produced the first of many published papers and regular updates to the Scientific Advisory Group for Emergencies (SAGE) and the four UK public health agencies. (8) Within 18 months, COG-UK had contributed one million genomic sequences taken from positive community tests around the UK, ultimately representing a quarter of all those collected globally by GISAID – thereby playing a major role in identifying new variants of concern throughout the pandemic. (9)
Only a few countries globally reached this impressive volume of genomic sequencing capability. However, since then, much of COG-UK’s sequencing network has been transferred to the newly created UK Health Security Agency (UKHSA), and the consortium officially closed in March 2023 with the website archived by the National Archives. (10)
Overcoming genomic sequencing challenges
Despite its immense potential, the widespread implementation of genomic sequencing still faces several challenges. One major obstacle is the lack of infrastructure and resources in low-and middle-income countries, which limits their ability to participate in global genomic surveillance efforts. Additionally, access to pathogen genomic sequences can be hampered due to many issues, for example, over rights of publication and credit sharing. (11) Investment in national and regional genomics infrastructure and facilities and ensuring interoperability between data repository systems should be a priority. (12)
Ways to facilitate this would be to promote international cooperation, collaboration, and exchange of good practice. Data protection and security of network and information systems must be robust so that confidence in genomic data sharing or, indeed, any cross-border exchange of patient-derived data of any kind is upheld to the highest levels of data control measures and regulation. Innovation in informatics and continuous improvement will be required to support the ever-increasing amounts of genomic data generated and used to inform the development of new and evolving care pathways.
Conclusion
Few technologies are as transformative as genomics. It contributes to our understanding of pathogens’ origin and transmission patterns, monitoring their evolution, improving diagnostic capabilities, and guiding intervention strategies. Genomic sequencing and data analysis are crucial in pandemic preparedness and prevention. By fostering collaboration and promoting data sharing, scientists can harness the power of genomics to mitigate the impact of pandemics, ultimately saving lives and safeguarding public health.
References
- https://www.nejm.org/doi/full/10.1056/NEJMp230508
- https://www.ovg.ox.ac.uk/news/oxford-scientists-how-we-developed-our-covid-19-vaccine-in-record-time
- https://md.catapult.org.uk/lighthouse-lab/
- https://www.gov.uk/government/publications/technical-report-on-the-covid-19-pandemic-in-the-uk/chapter-6-testing#technologies
- https://covid19.who.int/
- https://gisaid.org/about-us/history/
- https://webarchive.nationalarchives.gov.uk/ukgwa/20230505083137/https://www.cogconsortium.uk/
- https://www.bmj.com/content/381/bmj.p115
- https://www.bmj.com/content/381/bmj.p1157
- https://www.cogconsortium.uk/
- https://www.nejm.org/doi/pdf/10.1056/NEJMp2304214?articleTools=true12

This work is licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International.