Here, several experts from the International Society for Biological and Environmental Repositories provide a novel perspective around distributive biobanking models, including why biospecimens need blockchain
When you are sick, to determine what is wrong with you, a doctor will likely require you to give up a piece of your body (e.g. tissue biopsy, blood samples), some secretions (saliva, cervical swab) or some of its excretions (e.g. urine, faeces). They do this as ‘biospecimens’ are central to their understanding of human biology and the diseases we encounter.
At its core, ‘biospecimens’ contain the information that doctors and researchers use to understand what type of disease a patient has and to determine possible treatment options that should be used for their clinical management. Through advances in biotechnology, the information in the biospecimen can now be readily and routinely extracted leading to vast amounts of digital data being captured in machine-readable forms. This ‘datafication’ of biospecimens generates different types of ‘omic’ data (genomics, transcriptomics, proteomics, metabolomics, epigenomics) reflecting the molecular events ongoing in the disease. Or it may be information drawn from medical and microscopic analyses of images of tissue samples. When building a framework for personalised treatment of disease, the complexity of the biospecimen-derived information must be captured in meaningful and actionable ways. As the impacts of the use of digital information to the healthcare system have been emerging, personalised medicine will be directed by computational scientists and data analysts active in the biomedical domains. In doing so, tissue-based science is now an informatics problem.
The problem of centralising biospecimens
Within biomedical research, centralisation of its core activities into aggregated facilities or ‘repositories’, such as nodes, databases, or registries is a well-entrenched practice. Current tissue handling practices seek to aggregate biospecimens into giant freezers that are linked to databases of annotated clinical and ‘omic’ data, registries of patient information and research project ethical consent.(1) Over the past 20 years, aggregation of biospecimens into biobanks have emerged which, in the U.S., is anticipated to become a $2.7 billion industry by 2022.(2) Biobankers operate their repositories as largely autonomous self-governed entities that obtain their biospecimens through arrangements within pathology services or directly from surgery. Whilst the narrative around strategic planning recognises that ‘biobanks are crucial for medical research,’(3) a bevy of reports highlight how low biobank utilisation fuels unsustainable biobank models.(4) Despite grand plans to aggregate biospecimens into ever-larger centralised facilities, they have not yet realised their potential. The reason for this is multifaceted and influenced by the following issues: parochialism in research, biohoarding of specimens without release to researchers(5), capacity issues, poor business models, separation of biospecimen from the patient record and disengagement of research from the healthcare environment.
Centralised biobanking suffers from other disadvantages known to plague centralisation models. Specifically, biospecimen aggregation creates single points of failure within the system which require excessive protections and constant vigilance to maintain security. Centralised storage of biospecimen without clear motivations for their successful use leads to legacy collections that have no purpose, leaving unused biospecimen-derived information, creating noise and raising the question of relevance. Finally, centralisation removes the biospecimens from their primary source, which is the patients themselves. This led to a loss of engagement of key stakeholders. The decision-making process reverts to an independent ‘authority’ who determines access rights, requiring donors to have unconditional trust in biobank practices and motives. In summary, aggregation of biospecimens into central facilities cacoons the information they could provide into systems that are controlled by a limited few who may censor the message biospecimens hold and thus dictate the research narrative whilst restricting open discovery and exploration by many others.
The decentralisation of biospecimens using blockchain
Bold and innovative strategies are needed for biospecimen derived informatics to impact digital health initiatives. Shifting aggregative biobanking to a model that manages biospecimens as an integral part of a digital health information flow will revolutionise how we learn from biospecimen-derived information. Distributed models rely on the presence of many independent participants that manage processes together through common standards performed in parallel. For distributed models to work, all components required for the system must be decentralised, including consent practices, ethics and governance oversight, patient engagement, biospecimen transfer logistics, datafication processes, quality management, research results dissemination and even clinical decision making.(6) Decentralised environments see processes distributed away from a central authoritative organisation or group, with decision making falling to a cooperative of people having different perspectives, leading to greater objectivity.
We envisage biobanks would benefit from blockchain technology. Blockchain as an incorruptible shared digital ledger allows for the distributed, secure, transparent and robust transactions by tracking the secure, ethical transfer of biospecimens to researchers. This blockchain implementation was created to build trustworthy decentralised applications that run with no downtime, censorship, fraud or third-party interference which is ideal in the context of exploring human tissue within the research context. Use of smart contracts has been identified as enabling the management for human subject regulation allowing Human Research Ethics Committees and governance bodies to comprehensively, transparently, securely and automatically administer the requirements for research integrity whilst avoiding repetitive intervention between participants.(7) Blockchain is now being used to personalise biospecimen collection and distribution allowing donors to determine how their specimens will be used in research (Figure 1). Blockchain technology enables ways of synchronising data between participants within a system that is not influenced by suspicion. If tissue-based science is an informatics problem, then biospecimens should be the first block in the chain of information flow.
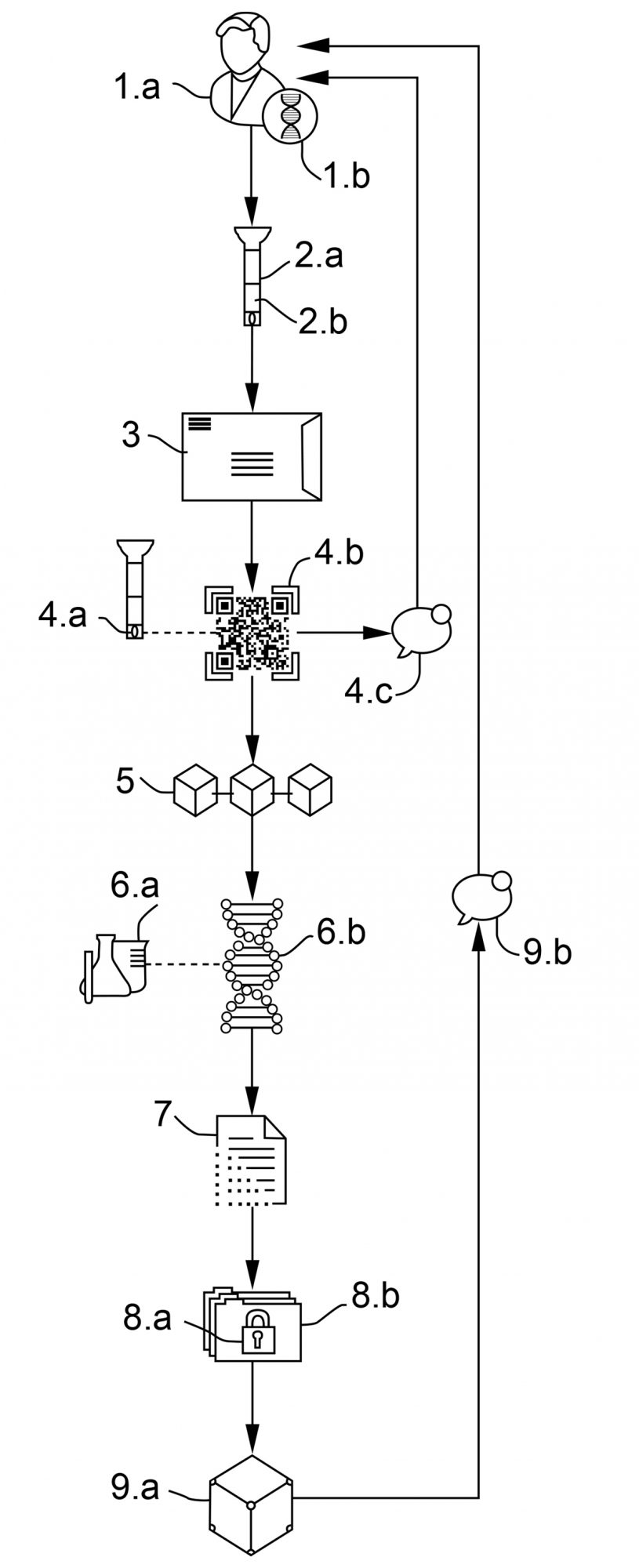
Figure 1: Proposed steps in a biospecimen- informatics blockchain developed by Genobank.io to tokenize biospecimens and its corresponding DNA/RNA datasets.
- (a) The donor creates a “BioWallet” (Genobank’s account) having a public address and private key. (b) The wallet encrypts user’s private key with a pin code provided by the user & creates an empty unique digital repository for the coming “multi-omics” data sets.
- (a) The donor deposits their biospecimen into a QR/Barcoded tube. (b) The donor scans the tube code creating a unique non-fungible token (NFT) in their biowallet containing information including GPS location, expiration dates, brand, date and time recorded as metadata.
- The donor ships the biospecimen to an assigned biobank or laboratory.
- (a) The biobank receives the biospecimien, scans the QR/Barcode and connects via internet to genobank.io (b) Each biorepository/tube is referenced to a specific BioWal- let as a token where the owner of the biowallet is the only one that can receive, decrypt & share the corresponding data sets and messages. (c) Once the biosample is received by biobank, the donor receives a text notification or tokenized reward.
- The transactions are recorded in a public blockchain specifying the use of the data criteria.
- (a) Biobank/laboratory process the biosample and applies all the amplification or preparation libraries. (b) The DNA/RNA is extracted and the process digitised.
- The DNA/RNA is sequenced using the best available technology.
- (a) The digital raw data is generated (b) Raw data is encrypted using donor’s private key and a non-fungible token is created to claim ownership of this data sets in the blockchain. Fungilble tokens can be created from this data sets to allow releasing serialised copies using the corresponding biowallet where the private key must be used to sign all the transactions.
- (a) Encrypted raw data is stored in the assigned private decentralised digital repository (b) Donor is notified that their data is available through the encrypted private messaging system.
References
- Richard E Gliklich, Nancy A Dreyer, “Registries for Evaluating Patient Outcomes: A User’s Guide”, 3rd edition Editor: Michelle B Leavy, Rockville (MD): Agency for Healthcare Research and Quality (U.S.), 2014.
- https://www.marketsandmarkets.com/PressReleases/biobanking-devices.asp
- Zhou L., Catchpoole D.R., “Spanning the genomics era: The vital role of a single institution biorepository for rare disease research over a decade.”, Translational Paediatrics, 4(2), 93-106, 2015.
- Cadigan RJ, Juengst E, Davis A, Henderson G., “Underutilization of specimens in biobanks: an ethical as well as a practical concern?” Genet Medicine, 16(10):738-40, 2014.
- D.R. Catchpoole, “Biohoarding: Treasures not seen, stories not told.”, Journal of Health Services and Research Policy, 21(2), 140-142, 2016.
- Kravchenko P., Skriabin B., Dubinina O., “Blockchain and Decentralized Systems.”, Volume 1, Publisher: Distributed Lab, Kharkiv, Ukraine, 2018.
- Olivia Choudhury, Hillol Sarker, Nolan Rudolph, Morgan Foreman, Nicholas Fay, Murtaza Dhuliawala, Issa Sylla, Noor Fairoza, Amar K Das. “Enforcing human subject regulations using blockchain and smart contracts.”, Blockchain in Healthcare Today, 1, 2018.
Daniel R. Catchpoole
Head, Tumour Bank, The Children’s Cancer Research Unit
Kids Research, The Children’s Hospital at Westmead, NSW, Australia
daniel.catchpoole@health.nsw.gov.au
Daniel R. Catchpoole & Paul Kennedy
School of Computer Science, Faculty of Engineering and Information
Technology, The University of Technology Sydney, Sydney, Australia.
www.uts.edu.au/about/faculty-engineering-and-information-technology
www.uts.edu.au/staff/daniel.catchpoole
https://paul-kennedy.weebly.com/
Daniel Uribe
Genobank, San Francisco, California, U.S.
Discussion Think tank
International Society for Biological and
Environmental Repositories (ISBER)
Tel: +1 604 484 5693