Assistant Professor Clare Kirkpatrick shares insight into the development of new antibiotics for the opportunistic human pathogen, Pseudomonas aeruginosa
From time to time, the same sensationalist phrases about the problem of antibiotic resistance appear in the news headlines: for example “ticking time bomb,” “antibiotic apocalypse,” “global threat,” “return to the dark ages of medicine” and so on. But the headline writers do have a point: many bacteria with a significant disease burden in humans are developing resistance to many drugs in clinical use and antibiotic resistance genes are spreading. Meanwhile, the number of existing molecules that have anti-bacterial activity, and the number of bacterial macromolecules (structural proteins, cell wall components, enzymes) which are targeted by these molecules, remains low. As the development of new antibiotics is unlikely to ever be a source of significant revenue for drug companies, very little industrial resources have been focused on it and the rate at which new antibiotics come to the clinic is very low. Discovery of new pathways within bacteria that could provide a source of new drug targets, as well as new molecules (or alternatively, existing antibiotics in novel combinations) that interfere with them, is a challenge that can more easily be met in an academic setting without the requirement to generate profits for shareholders.
Pseudomonas aeruginosa
I am currently establishing my research group at the Department of Biochemistry and Molecular Biology, University of Southern Denmark, with the aim of studying cell cycle and motility control in the opportunistic human pathogen, Pseudomonas aeruginosa. We focus particularly on how the factors controlling cell division and motility (bacterial movement) interact with each other and their potential as eventual drug targets. Pseudomonas aeruginosa is highly versatile in terms of the environments it can colonise, and can establish acute or chronic infections in burn wounds, skin ulcers implanted medical devices like catheters or pacemakers and the lungs of cystic fibrosis patients. It is also highly naturally resistant to many classes of antibiotics because it possesses multiple defence mechanisms against them. New drugs active against this species are therefore needed urgently. Indeed, carbapenem-resistant Pseudomonas aeruginosa was named by the World Health Organization in February 2017 at the top of the “Priority 1: Critical” category of the WHO Priority Pathogens List of bacterial species for which new drugs ought to be developed as a matter of urgency.
The ability of Pseudomonas aeruginosa to establish infections fundamentally depends on its ability to move around, in liquid or on surfaces, to find the best possible niche to proliferate. This movement is dependent on the flagellum, a corkscrew-like structure that protrudes from one end of the cell and works like a propeller to enable the cells to swim, and thread-like strands of protein called pili that extend and retract, allowing the cells to pull themselves along on surfaces. Both these structures are also necessary for the first stages of forming a biofilm, where the cells attach themselves to the surface they are going to colonise. The flagella and pili are always located at the ends of the rod-shaped Pseudomonas cell and this location is vital for them to function correctly. Mutants which produce flagella or pili out of the sides of the cell can no longer move properly and are therefore at a disadvantage for establishing a biofilm or colonising an infection site. Production of flagella and pili in the correct place is co-ordinated with the cell cycle and the environment that the cells are in, but the mechanisms that control it are not fully understood. In order to shed some light on this subject, my group is investigating the roles and interactions of the protein factors that control the location of flagella and pili using a chemical genetic approach.
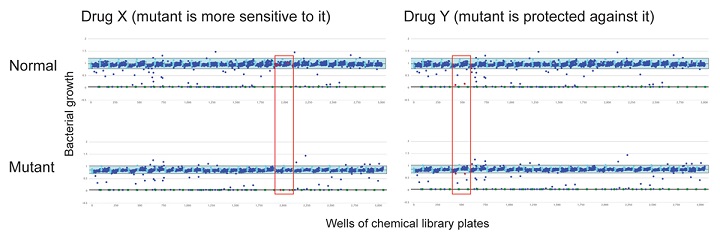
Bacterial Chemical genetics
Bacterial Chemical genetics is a relatively new research field in which drugs, chemicals, or other small molecules are tested against bacteria, not necessarily to see whether they have antibiotic activity or not, but to see which (if any) processes in the cell they interact with. This can be discovered by comparing the response of a normal, non-mutant strain and a mutant that is lacking one of the genes involved in the process of interest, to a collection of drugs or small molecules and identifying ones where the mutant responds differently (either by growing better in the presence of the drug, or worse) to the non-mutant strain. Both these responses indicate that the drug interacts with the process of interest, and can be followed up by using that drug in other experiments to find out what exactly it is interacting with. Work from several other laboratories has shown that this approach can identify missing enzymes in metabolic pathways or previously unknown factors involved in, for example, cell wall synthesis.
My group’s objective is to use the same approach to identify previously unknown factors involved in the control of flagella and pili positioning and function in Pseudomonas and to define what potential these factors have as drug targets. Our preliminary data on screening a panel of five Pseudomonas mutant strains lacking known control factors involved in this process against a 1280-compound chemical collection has identified 4 drugs with differential effects against one or some of the mutant strains (two examples are shown in the figure) and we are currently investigating their mode of action and the reason for the mutant-specific effects on growth. We were very surprised to see that in a few cases, instead of the mutations causing increased sensitivity to one of the compounds, they appeared to protect against it, despite the gene in question having no apparent relationship to the known function of the drug. Since this could indicate a previously unknown potential mechanism for antibiotic resistance, this interaction is a priority for my group’s short-term future work. In the long term, we hope to characterise how factors controlling Pseudomonas flagella/pili positioning and function are coordinated into a regulatory network and to understand which nodes in the network are the most critical for the cell’s ability to cause disease (and which could, therefore, be the best targets for novel antimicrobial compounds).
Clare Kirkpatrick
Assistant Professor
Department of Biochemistry and Molecular Biology, University of Southern Denmark
Tel: +45 6550 9094
clarek@bmb.sdu.dk
www.sdu.dk/bmb
Please note: this is a commercial profile