The Institute of Hygiene, Westfälische Wilhelms-Universität Münster highlights the changes in microbal pathogenicity
In many cases microbial pathogenicity cannot be predicted directly from the properties of the microorganism. Instead, often complex interactions between the environment and host, the host and microbe, and the microbe and environment have to be taken into account.
Today’s view on microbial pathogenicity has profoundly changed. Pathogenicity is no longer considered a specific property of the microbe, but thought to arise from a complex and dynamic interaction with the host. Microbes may grow and multiply only in certain hosts. Likewise, a pathogen might simply exist within a host and cause no signs or symptoms of infection or elicit disease to variable degrees, ranging from mild to deadly. Accordingly, we have to shift the focus onto the host-microbe interplay as a complex and malleable phenomenon governed by the properties of both the microbe and its host environment. To avoid a microbe or host-centred view and to rather implement a holistic approach, infectious disease-related research should be cross-disciplinary and integrate traditionally separated biomedical and life science disciplines. In order to open up new therapeutic avenues to interfere with infection, we have to explore bacterial pathogenicity factors and infection pathways, but also the role of co-colonising bacteria, abiotic and bioactive factors, and pathogen and host variables.
Furthermore, for many microbes, factors required for microbial pathogenicity in a normal host cannot be identified. Many pathogenicity-associated traits can be found in pathogenic, as well as in non-pathogenic variants of a species. Additionally, properties conferring pathogenicity depend as much on the host as they do on the microorganism. Recent developments in modern medicine further corroborate the fact that in many cases pathogens and non-pathogens cannot be unequivocally distinguished based on specific traits, because the introduction of broad-spectrum antimicrobials, immunosuppressive therapies, modern treatment options (including, organ transplantation and joint replacement, implantable devices and indwelling catheters), each of which alters host-microbe interactions, created conditions, in which the host became susceptible to microbes that were previously considered non-pathogenic. Consequently, many microbes previously considered non- or facultative pathogenic, such as certain Escherichia coli variants, Pseudomonas aeruginosa, Staphylococcus epidermidis, fungi (e.g. Aspergillus fumigatus, Candida albicans), turned out to be able to cause serious disease.
Many microbes cause disease in some, but not all of those individuals who are colonised by them. This is exemplified by bacteria such as staphylococci, which are actually present in most individuals, but only cause disease in some. There is also growing evidence from microbiome analyses, that alterations of the normal composition of the microbiota or of the balanced relationship between microbes and the host barriers, can making the host more susceptible to infection by microbes that usually do not cause disease. Pathogenicity is therefore an outcome of host-microbe interaction and is thus linked to characteristics of the host, as well as those of the colonising and infecting microbiota.
Continual research to monitor microbial pathogenicity
Despite the ongoing research activities worldwide regarding bacterial pathogenesis and the development of antibiotic resistance and pathogenicity-associated traits, the underlying evolutionary mechanisms and genetic elements in relation to the complex interactions between pathogens and their hosts are not yet well understood. We study bacterial genome plasticity and gene expression in vivo to understand bacterial interaction with their hosts during infection or colonisation which may result in differential susceptibility to infection (Figure 1). Next-generation sequencing (NGS) allows the comprehensive genome wide identification of DNA sequence alterations, such as re-arrangements, insertion/deletion as well as horizontal gene transfer events. The use of (meta-) genomic data and NGS tools facilitates the analysis of genomic alterations in vivo as well as the analysis of bacterium-host interaction at the transcriptional level by dual transcriptome sequencing. An increased understanding of bacterial genome plasticity and gene expression in vivo will support the development of preventive, therapeutic and diagnostic strategies to specifically interfere with relevant virulence or colonisation mechanisms of pathogens in the reservoir and the infected patient.
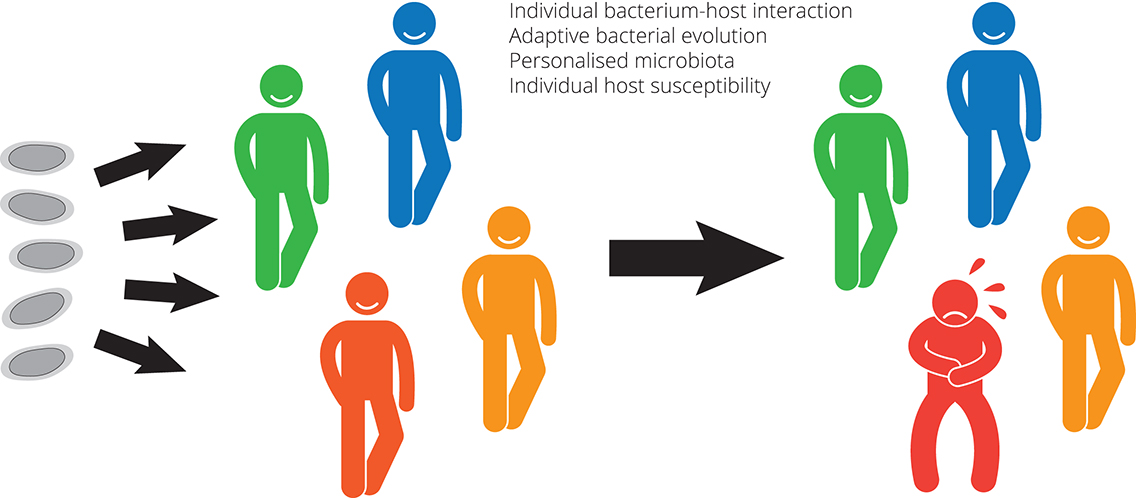
Bacterium-host interactions drive adaptive changes in bacteria, their phenotypic diversity and genome complexity. Escherichia coli (E. coli) is a versatile pathogen, acting as a major cause of morbidity and mortality as well as a commensal, which colonises the human gastrointestinal tract within hours after birth, and typically builds a mutualistic relation. We analyse genome plasticity and gene expression in vivo and the mechanisms by which E. coli adapts during long-term colonisation of the urinary tract or the intestinal tract of individual human hosts. Long-term colonisation of the urinary bladder or the intestinal tract by individual E. coli strains is one therapeutic strategy to counteract infection by bacterial pathogens in these niches. In addition, isolates of uropathogenic E. coli from different time points of a chronic urinary tract infection also allow to monitor bacterial adaptation during infection.
Comparative and functional genomic analysis of E. coli re-isolates from the bladder of deliberately colonised patients or re-isolates from the intestine of individuals colonised with an E. coli strain identified unique bacterial adaptation patterns in each person supporting the idea that, in addition to stochastic events, adaptive bacterial evolution is driven by individual host environments. Our data support the idea that hosts appear to personalise their microbiota. Furthermore, our studies of pathogenic and non-pathogenic, commensal E. coli isolates have identified that, rather than conventional pathogenicity factors, determinants involved in bacterial fitness, e.g. in nutrient uptake and utilisation, as well as protection against the host’s immune response are advantageous as they allow rapid adaptation and growth of these strains in niches where they can cause infection. We are convinced that the combined analysis of bacterial and host responses during infection will define promising targets for novel antimicrobial strategies. Defining a role for metabolic traits during disease and identification of novel processes underpinning this capability could allow development as potential approaches (e.g. dietary manipulation, attenuated vaccines or anti-infectives) for preventing infection.
Prof. Dr. Ulrich Dobrindt
Institute of Hygiene
Westfälische Wilhelms-Universität Münster
dobrindt@uni-muenster.de
https://campus.uni-muenster.de/en/hygiene/research/research-topics/microbial-genome-plasticity/
Please note: this is a commercial profile