Professor Darren Griffin and Dr Rebecca O’Connor from the University of Kent’s School of Biosciences, discuss their work on improving UK pig production and reaching out to South East Asia
The domestic pig provides 43% of meat consumed worldwide and is, thus, the leading source of meat protein globally. In most countries, commercial pig breeding operates in a pyramidal structure with the top tier (nucleus level) consisting of purebred boars that are selected on their genetic merit (meat quality, for instance, is an indicator). One aspect however that is sometimes given less priority is the fertility of the boar.
Any fertility problems in these top (nucleus) boars can have a profound effect on litter sizes and fecundity rates. This ultimately leads to a reduction in food production and higher environmental costs, as unproductive males produce unnecessary greenhouse gases. When a fertility issue exists, it is often caused by a chromosome rearrangement (the exchange or ‘translocation’ of parts of the genome) which in males, will result in a high proportion of genetically unbalanced sperm. In boars, a higher number of unbalanced sperm will result in significantly smaller litter sizes – between 25% and 50% smaller than expected.
Artificial Intelligence (AI)
These problems are perpetuated further through an increasing emphasis on artificial insemination (AI). Currently, when AI is used across the breeding population, the male to female ratios are 1:1000, therefore, the potential financial and environmental costs of using an infertile boar in the AI breeding pyramid are enormous. The need, therefore, to identify animals that carry a translocation and prevent their use in the breeding population is paramount. Indeed, pig semen (because of the quality of its genetics) is a major export for the UK economy.
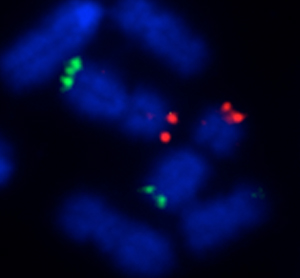
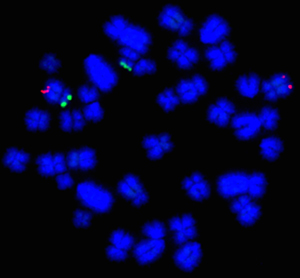
The presence of a chromosome translocation cannot be identified through routine semen analysis as affected boars look perfectly normal. The industry-standard approach to screening for these abnormalities has been to use a method known as ‘karyotyping.’ Now routinely used for translocation screening, many breeding companies karyotype to proactively screen their boars before entry into the AI stud. In fact, over the last 40 years, ~170 different chromosome translocations have been identified (across all breeds) using this method with a reported incidence rate of 1/200 (0.5%) of boars used for AI. Indeed, in France, it is a legal requirement that all boars used for AI are certified as translocation free.
CytoScreen Solutions
However, karyotyping is labour-intensive, it requires pig-specific expertise and does not allow identification of the very smallest (‘cryptic’) translocations. To overcome these drawbacks, here at the University of Kent, we have established a new service called ‘CytoScreen Solutions’. At the core of this service, is a method we have developed to solve the inherent problems of karyotyping in a rapid, accurate and cost-effective manner. We use an innovative adaptation of a technique known as ‘FISH’ (fluorescence in situ hybridisation). With this new method, we can identify even the smallest of translocations (as demonstrated in the figure).
To date, CytoScreen Solutions has screened over 1,600 boars with this technology and has found over 70 (5%) with translocations. Our results strongly suggest that translocations in the breeding population are significantly higher than previously thought. In addition to our in-house screening service for pigs, we are actively expanding to North America, Australia and South East Asia, where our method has now been used in labs in all three continents. One of our most recent projects has been to use this University of Kent technology in Thailand to make pig production more cost-effective and environmentally sustainable. Pig breeding is a major and growing industry in South East Asia and our goals here are to support local teams in their improvement of the security and resilience of the pig breeding industry.
The CytoScreen Solutions method will support initiatives in South East Asia to improve the genetic quality of their breeding stock, thereby ensuring long-term market growth and reducing the financial and environmental risks associated with excess waste. Improved margins for the pig breeding industry will ensure profits can be re-invested in local businesses, ensuring long-term growth and sustainability of this market. Importantly, this improved investment will also have the benefit of ensuring that the emphasis is on generating high-quality meat products, bred under first-class welfare and environmental conditions.
The improved profitability provided through this investment will allow the breeding industry to expand, supporting local jobs and ensuring sustainable economic growth both for industry leaders and workers in local communities. Passing on these savings to the end-consumer will have the consequence of improving nutrition and will reduce reliance on cheaper imports, ensuring that local suppliers benefit from gains in genetic progress developed in the UK and Europe.
In the broader sense, screening for translocations using the highly accurate CytoScreen Solutions method will ultimately result in a more efficient food production chain, with an emphasis on generating nutritious, high protein food for a wider population. As described, these benefits are not just limited to well-established breeding industries, such as those in Europe, but can be adopted by communities worldwide. While the reduction in cost due to improvements in efficiency are of interest to both producers and consumers, the environmental and animal welfare benefits of reducing waste are an increasingly important benefit to all.
For more information, please contact Dr Rebecca O’Connor at r.o’connor@kent.ac.uk.
O’Connor, R. E. et al. Isolation of subtelomeric sequences of porcine chromosomes for translocation screening reveals errors in the pig genome assembly. Anim. Genet. 48, (2017).
Please note: This is a commercial profile











