Frederique Lisacek from SIB Swiss Institute of Bioinformatics, provides the computer-based tools for exploring ways of breaking the glycocode
Over the past two decades, the word “genome” has increasingly become part of the usual conversation and the SARS-Cov2 pandemic has sealed this popularity. The “ome” suffix appended to many common terms of molecular biology reflects researchers’ concern for comprehensiveness in so-called “high-throughput” or “large-scale” studies. Corresponding “omics” methods get scientists closer to full coverage of a research topic. In this landscape, a genome cannot be solely considered and its analysis initiates the next steps in understanding functional aspects of life, one of which involves the “proteome”.
A look at proteins
Proteins – encoded in the genome – are the effectors of cellular processes. They adopt very specific shapes to perform very specific functions. In 2021, the Protein Data Bank (PDB), the universal repository for protein three-dimensional structures celebrates its 50th anniversary. The PDB is one of the landmarks of bioinformatics, the computational pillar of research in molecular biology and particularly prevalent in “omics” studies where biological data analysis is heavily automated.
High throughput protein science has achieved progress through mapping molecular interactions to elucidate protein assembly into large functional complexes or as part of processes of recognition, signalling, etc. (1) However, not all protein structures have been solved and the repertoire of structural changes modulating protein function(s) is yet to be fully established. These structural changes have been reported to be induced by mutations, but less commonly through the action of enzymes that will modify a component (amino acid) in the protein. Living processes are full of nested situations and it should be noted that enzymes are proteins that also often need to be modified by themselves or other enzymes. Understanding these so-called “posttranslational (2) modifications” (PTMs) is a major challenge of protein science.
A protein is synthesised as a chain of a few hundred amino acids – on average – then folds into three dimensions. Modifying enzymes act on a few to dozens of sites on the protein. Some modifications are constant such as the addition of a phosphate group which plays a key role in channelling information in and out of the cell. Others are variable such as the attachment of carbohydrate molecules composed of a variety of chemical groups. Our team is specialised in designing and providing the bioinformatics means to process and interpret experimental results that contain modified proteins, especially those with complex carbohydrate molecules (also called glycans) attached. (3)
Glycoproteins and viruses
The term “glycoprotein” is frequent in biology textbooks or in research articles though with no or little reference to the attached carbohydrates. It mostly spans the subcategory of proteins active at the cell surface. For example, viral surface proteins are often named “gp-xxx” such as gp120 of HIV where “gp” is short for glycoprotein. In 2003, the “glycan shield” expression was coined to depict the high density of carbohydrates detected on the gp160 of HIV-1. On the host side, viral receptors were shown to bind viral glycans thereby playing a crucial role in interspecies transmission.
The fine specificity for alike, but distinct glycans was highlighted to differentiate avian from human flu. Many enveloped viruses deploy a glycan shield, but common representations do not account for this property. Yet, 2020 may be a turning point. The COVID-19 pandemic and three Nobel prizes in Medicine have provided a unique opportunity to highlight the importance of glycosylation and more researchers are now picturing “hairy” as opposed to smooth SARS-Cov2 spike proteins. (4) With the SARS-Cov2 data deluge, it has become impossible to ignore the striking presence of glycans that decorate the protruding surface protein.
Even though the vast majority of virologists focus on viral genomic sequences, glycobiologists have long considered glycan-virus interactions since viruses hook onto host glycoconjugates (glycoproteins as well as glycolipids) to enter cells.
These glycan -mediated interactions involve a variety of host receptors, as well as viral surface proteins known as hemagglutinin in influenza or spike proteins in rotaviruses and coronaviruses. In fact, glycan-binding has been under scrutiny for decades to elucidate the role of two main categories, i.e., blood group antigens (blood group molecules are glycans) and sialic acids (common constituents of glycans). For example, it was shown that different strains of noroviruses, causing gastroenteritis, have distinct preferences for human O or A blood-group phenotypes. These studies confirm that virus glycosylation should be considered in vaccine design to trigger the antibody production that targets protein regions less likely to evolve through resistance mechanisms.
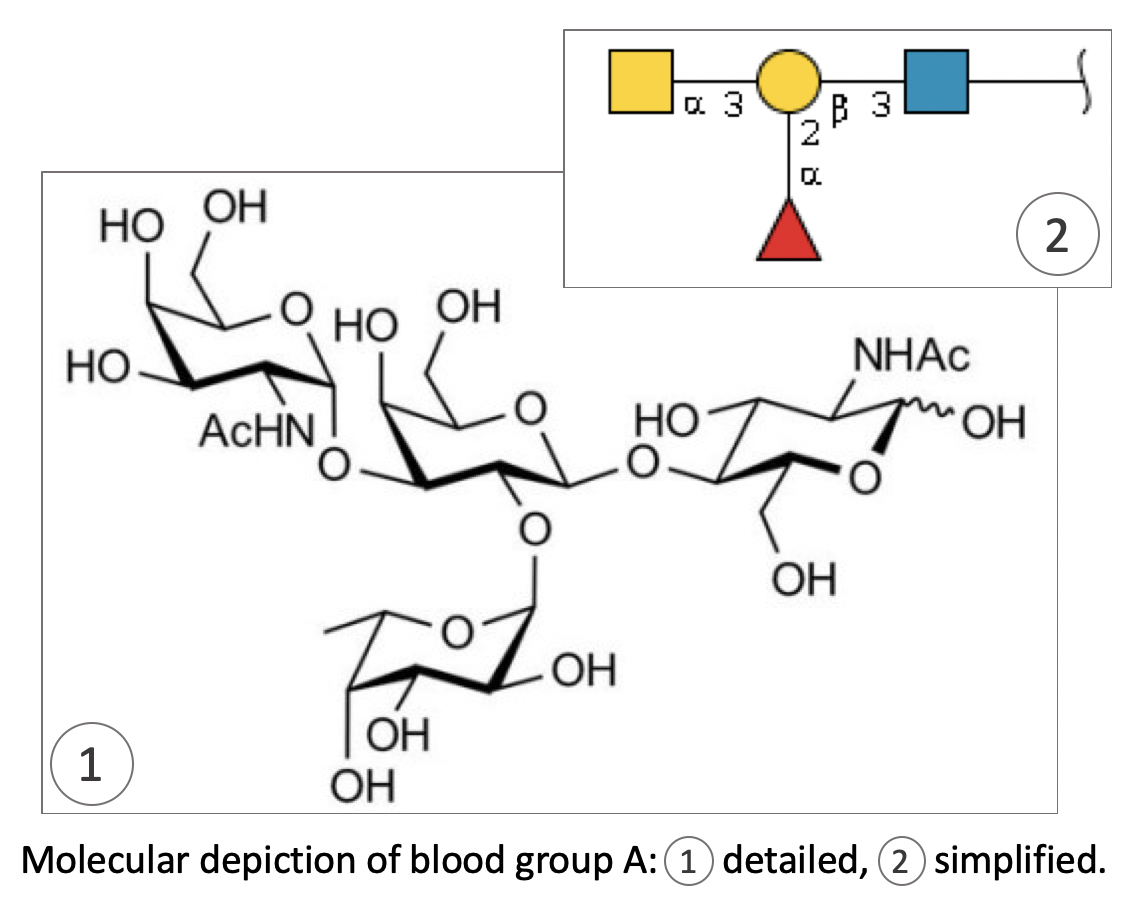
Overall, the situation as depicted so far emphasises the need for collecting data and recording glyco-related information to fully characterise viral infection. We undertook this task not only to support virology applications, but in the broader context of understanding the role of glycosylation. We provide a range of glyco-related databases and software tools that are available in the “glycomics” section of the portal of the SIB Swiss Institute of Bioinformatics called Expasy. With these, researchers with little or, indeed, extended knowledge of glycobiology can explore, search and compare glycans and glycoproteins in GlyConnect, our main platform for studying glycosylation.
The PTM code & the glyco-code
As mentioned earlier in this article, the next frontier in understanding protein function is deciphering the rules that govern the combinatorial occurrence of post-translational modifications, aka the “PTM code”. Glycosylation as the most variable of such modifications complicates the picture with its own challenging “glyco-code” borne by surface glycoconjugates or microbial membrane polysaccharides. The last category of molecular actors of this dynamic picture of the cell surface are the glycan-binding proteins known as “lectins”. This ability makes these proteins the key readers of information encoded by glycans. Lectins are diverse within and across all species nonetheless, they are poorly characterised in general-purpose protein databases.
“A protein is synthesised as a chain of a few hundred amino acids – on average – then folds into three dimensions. Modifying enzymes act on a few to dozens of sites on the protein.”
In 2019, we launched the UniLectin platform in collaboration with CERMAV in Grenoble, France, to address this issue based on a new lectin classification. This refined and more relevant description brings out lectin properties for an improved characterisation.
A community-based approach
Glycoscience is now reaching out to other omics following recent progress in carbohydrate structure resolution and synthesis as well as functional screening methods. The roles of glycans and glycoconjugates are manifold and revealed in various medical, biochemical and biotechnological applications. In recent years, roadmaps for glycoscience released in the U.S. and Europe have foreseen the growing need for centralised databases and associated software to match progress in high-throughput technology.
In the open access era, bioinformaticians strive to facilitate data sharing and information exchange. This approach applies to glycomics (glycoinformatics) through the GlySpace Alliance that gathers three complementing glycoscience portals developed in three continents. We represent Europe. This collaborative strategy is geared toward providing high quality, reliable, well-referenced and accurate data to the Life Science community. This collaborative strategy is geared toward providing high quality, reliable, well-referenced and accurate data to the Life Science community. Ultimately, it is intended to bring scientists closer and faster to breaking the glyco-code.
References
(1) See this video for an illustration.
(2) A protein is the result of translating a gene. Once this translation is over, any further molecular event involving that protein is qualified as posttranslational.
(3) Proteins with glycans are called “glycoproteins” (proteins with glycans) or “glycosylated proteins”.
(4) See New York Times illustration here.
Please note: This is a commercial profile
© 2019. This work is licensed under CC-BY-NC-ND.