Ute Deichmann of the Jacques Loeb Centre for the History and Philosophy of the Life Sciences at Ben-Gurion University, explores the role hierarchical causal models have on constancy and plasticity in biology
In natural history, notions of plasticity and change long antedated those of constancy and robustness. With his theory of the constancy of species, Linnaeus in the 18th century put an end to widespread notions of plasticity and transformation of species and thus laid the basis for a scientific understanding of species change.
Theories of the evolution of species as well as the germ theory of disease became scientifically meaningful only after the stability of organismic species sometime over long periods of time had been generally accepted. The idea of constancy became prevalent in many fields of biology in the late 19th century, especially in genetics, development and evolution, when constancy became inseparably linked with three basic biological principles:
- The structural and organisational hierarchy in organisms,
- Genetic causality of fundamental life processes,
- Biological and genetic specificity or genetic information.
These three principles are highlighted in particular in developmental biology, where the notion of constancy is prevalent, and where, in the words of Greg Gibson, “despite the fact that it takes 20.000 genes to make a complex multicellular organism and these have to work in very diverse environments, development works and leads to a constant outcome” (Gibson 2002; see Fig. 1).
Hierarchical gene regulatory networks in development
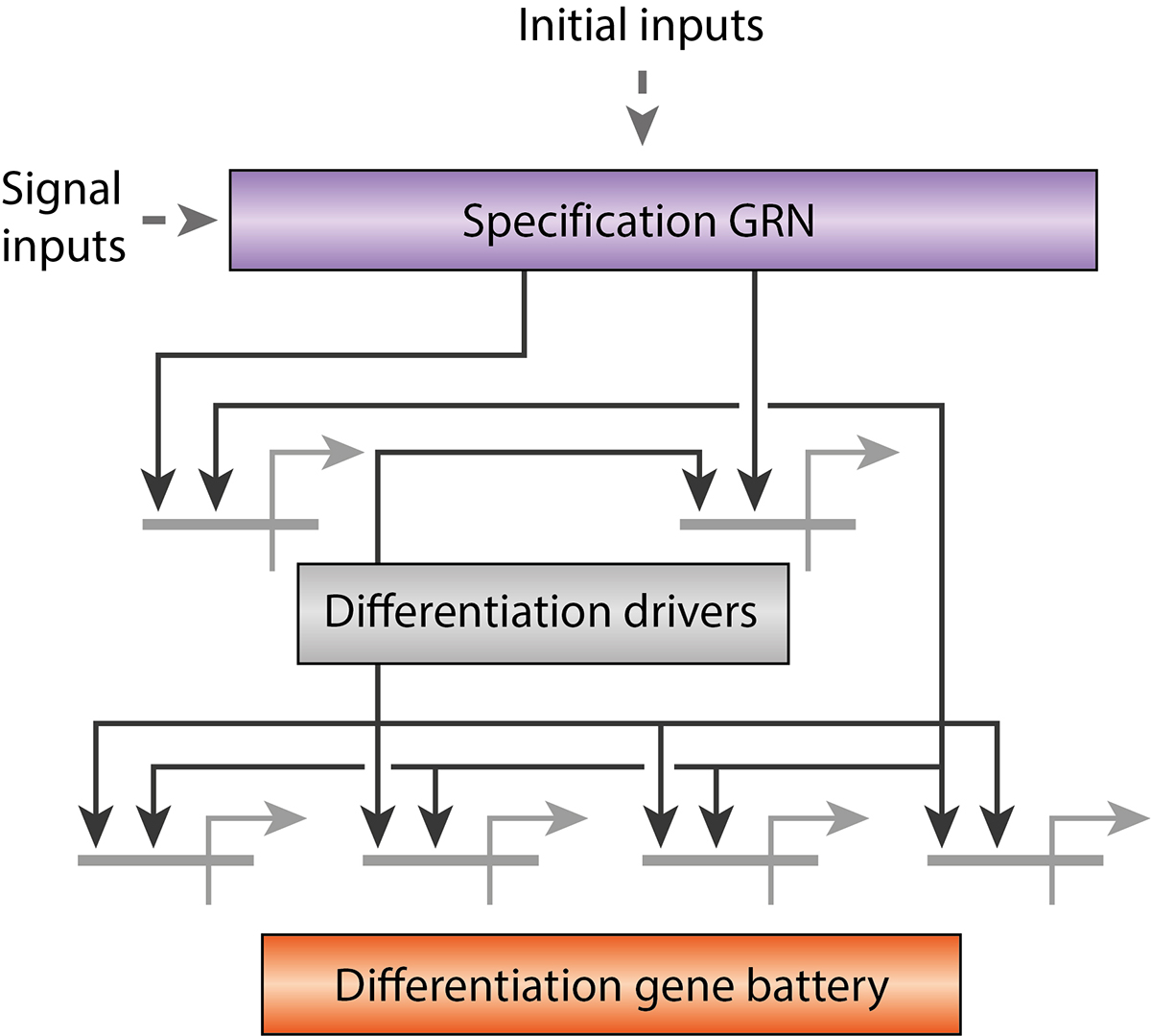
The constancy of animal development has been explained by hierarchical gene regulatory networks (GRNs) in which specific regulatory proteins, in particular transcription factors, play a major role (e.g. Davidson 2006). GRNs consist of regulatory genes and signaling pathways that execute a cascade of molecular mechanisms to transform an egg cell into a complex organism (Fig. 2). Davidson’s model has also implications for the theory of evolution: The most central genetic circuits of a GRN controlling development are so constrained that their variations are rare. This hypothesis explains the remarkable degree of constancy in evolution, i.e. the phenotypic stability of animal body plans that in some cases has persisted since around 500 million years ago.
Plasticity and unpredictability
Development is not only characterised by constancy and predictability, but there is also plasticity and unpredictability. The chemistry of life is characterised by molecular fluctuations and stochastic events in cells that seem to contradict deterministic explanations of development. The examination of the complicated hierarchy of buffering in cells and organisms to maintain constancy, e.g. through GRN and other mechanisms, is a fascinating challenge for current and future research.
The phenomenon of phenotypic plasticity, i.e. the generation of alternative phenotypes from the same genome, shows that not every single developmental trait is fully determined by particular genes. The limited effect of the environment on phenotypes was proposed already in the early 20th century by the Danish botanist Wilhelm Johannsen who equated the genotype with the notion of reaction norm, which referred to the range of potential – reversible – phenotypic variations in different environments. An intriguing example is the transition between solitarious and gregarious locusts elicited by mechanosensory input (Fig. 3).

Throughout the history of modern biology, the ideas of genetic causality and biological specificity have been rejected or marginalised in various fields. Around 100 years ago, the movement of biocolloidy, focusing on unspecific physical mechanisms replaced the search for relations of specific structures and functions by theories related to surface actions, electric charges and adsorption. All biochemically relevant substances of the cell such as proteins, enzymes and nucleic acids were regarded as biologically active colloidal aggregates of undetermined composition. The success of macromolecular chemistry and, subsequently, molecular biology, brought forward approaches that were able to causally explain the phenomena of biological specificity, rendering biocolloidy obsolete.
Questioning genetic causality
More recently, some approaches of epigenetics try to call into question genetic causality by claiming that small, unspecific molecules such as methyl groups are able to regulate gene expression. Social scientists and some biologists believe that these epigenetic marks are environmentally caused and can be inherited over many generations, thus marginalising the causal role of the genome for development. However, these approaches ignore established scientific facts in genetics and cell biology, according to which gene regulation is brought about by specific regulatory proteins. Because the enzymes that transfer epigenetic marks to DNA or histones lack DNA-sequence specificity, they require sequence specific factors such as transcription factors to guide their activity on the genome. Thus, the factors that are involved in gene regulation are hierarchically organised.
Likewise, current attempts to explain animal development by non-hierarchical, multilevel, multifactorial mechanisms, deny the relevance or even existence of causal relationships between specific regulatory factors. They reject the explanatory power of hierarchical GRN on the grounds that transcription factors contain intrinsically disordered (ID) protein regions that render them unsuitable for regulatory purposes. However, it has been shown that these ID-regions occur predominantly in domains that are used e.g. for recruiting co-factors, and less in the DNA-binding domain. The fact that ID regions contribute to the instability of transcription factors is an important pre-requisite for their suitability for regulatory functions.
Non-hierarchical, multilevel, multifactorial network models may explain phenomena of plasticity. But they do not convincingly explain how:
- Development can result in a functioning organisation,
- The development of individuals of a species always results in the same body plan, largely independently of the environment,
- How species can remain constant in different environments over long periods of time.
Historians and philosophers of science cannot predict scientific developments, as was formulated by biochemist and Nobel Laureate Otto Meyerhof some 90 years ago: “But one will only expect from scientific philosophy the consistent order of the system of scientific theories and no prediction of their contents.” However, historians and philosophers of science not only highlight the intellectual history of currently important concepts. They can also shed light on errors of reasoning and scientific dead ends, often due to neglect of basic biological principles that have been developed and revised since the late 19th century.
References:
Davidson, E.H. (2006). The Regulatory Genome. Gene Regulatory Networks in Development and Evolution. Burlington: Academic Press.
Gibson, G. (2002). Developmental Evolution: Getting Robust over Robustness. Current Biology 12, 347-349.
Deichmann, U. (2007). “Molecular” versus “Colloidal”: Controversies in Biology and Biochemistry, 1900–1940, Bulletin for the History of Chemistry 32, 105-118.
Deichmann, U. (2017). Hierarchy, Determinism, and Specificity in Theories of Development and Evolution. History and Philosophy of the Life Sciences 39 (4), 33. doi: 10.1007/s40656-017-0160-3
Deichmann, U. (2020). The Social Construction of the Social Epigenome and the Larger Biological Context. Epigenetics & Chromatin 13, 37. https://doi.org/10.1186/s13072-020-00360-w
Please note: This is a commercial profile