Dr Helen Rowe, Senior Lecturer in Epigenetics, outlines the significance of research into dark matter and genome remodelling, discussing the European Research Council funded TransposonsReprogram
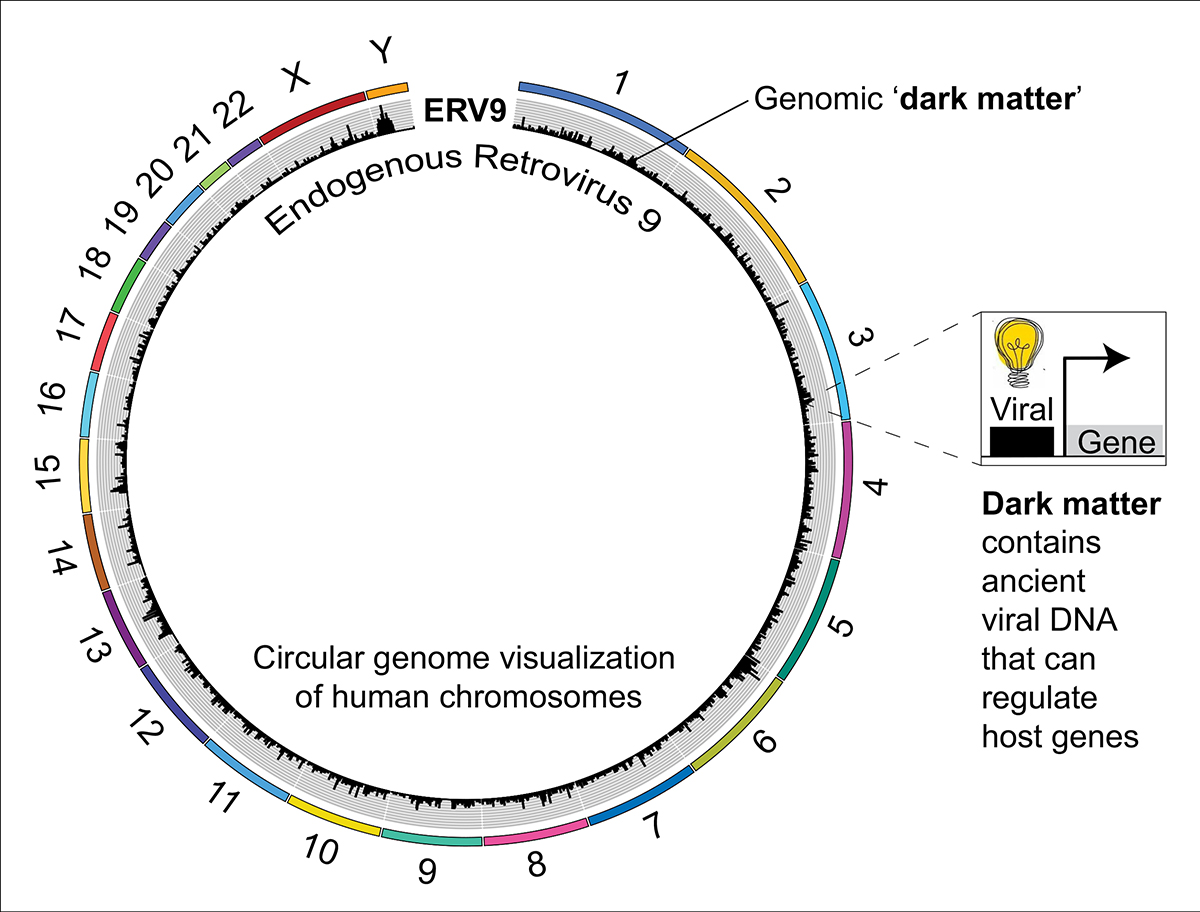
When the sequence of the human genome was released in 2001, it was revealed that protein-coding genes form less than 2% of our DNA. The nature and function of the remaining 98% is a cutting-edge area of active research and this less well-characterised portion of our genome is referred to as ‘genomic dark matter’.
What do we know about genomic dark matter and where did it come from?
It is estimated that up to 75% of the human genome is derived from transposable elements, which can be regarded as ancient viruses because of their ability to replicate. Most transposable elements replicate through a copy-and-paste method, which has allowed them to invade our genome and expand in number. This process is known as ‘transposition’ and leads to the insertion of extra pieces of DNA within our own chromosomes. When this occurs in the germline, these changes to our DNA will be inherited by our offspring, a prospect that sounds alarming, if not life threatening.
However, these events are extremely rare because although the bulk of our DNA is derived from transposable elements, the majority of these ancient viruses lost their ability to transpose millions of years ago and are now merely fossil remains.
The DNA of the human genome, therefore, reflects millions of years of genome invasions of now extinct ancient viruses, within which our protein-coding genes reside. Intriguingly, not all transposable elements are dead fossil remains. Several ‘master copies’ are intact and have retained the capacity to replicate and insert new copies of their DNA potentially into the middle of an essential host gene. It is only the LINE-1 (Long INterspersed Element) class of transposable elements that could still transpose or ‘jump’ within the human genome. To prevent this from happening, we have a multitude of host defence systems, particularly to protect our germline from mutations.
What does dark matter do?
As mentioned above, upwards of 50% of our genome is occupied by the fossil remains of ancient viruses, and these transposable elements have been referred to as ‘junk DNA’ because of their largely uncharacterised nature. However, transposable elements can control our own genes, and this ground-breaking discovery was made by Barbara McClintock in the 1940s. McClintock was a brilliant geneticist who not only discovered transposable elements, through her studies on maize, but she also realised that they could regulate host genes and she therefore described them as ‘controlling elements.’
How does dark matter control host genes?
This question comes back to the ‘viral’ nature of transposable elements: When they land in our genome, they in fact already have all the machinery necessary to regulate host genes. This is because, within a short stretch of DNA, they contain their own pro-moters, enhancers and repressors.
Recent research in this area has shed new light onto the breadth of functions of dark matter, which includes their roles in chromatin structure and nuclear organisation, as well as their participation in gene regulation. My own labs’ work and that of other labs has recently shown that LINE-1 nucleic acids are at the heart of mounting innate immune responses, which serve to defend ourselves from pathogens.
Can dark matter control cell fate?
A key question is: ‘Can dark matter control whole sets of host genes that define cell fate?’ Most examples of dark matter controlling host genes have been focused on a few isolated genes and well-studied types of transposable elements, such as endogenous retroviruses.
Our European Research Council funded work aims to understand whether dark matter can control whole gene expression programmes, which will, for example, allow a cell to transition to a different state. With traces of ancient viruses now scattered all over the human genome and capable of functioning as regulatory hubs, it’s easy to imagine how they could switch on or off whole sets of host genes simultaneously.
Another of Barbara McClintock’s discoveries was that transposable elements exist in different ‘families’, which are independent of each other. This is because these families reflect genome invasions and expansions by different classes of transposable elements over evolutionary time. Because different copies from the same family have similar or identical DNA sequences, they could all be targeted at once by the same transcription factor, explaining how an entire transposable element family network might be switched on. If they form a network of enhancers, for example, they could then switch on a whole network of nearby host genes through cis-regulation.
Cell fate and genome remodelling
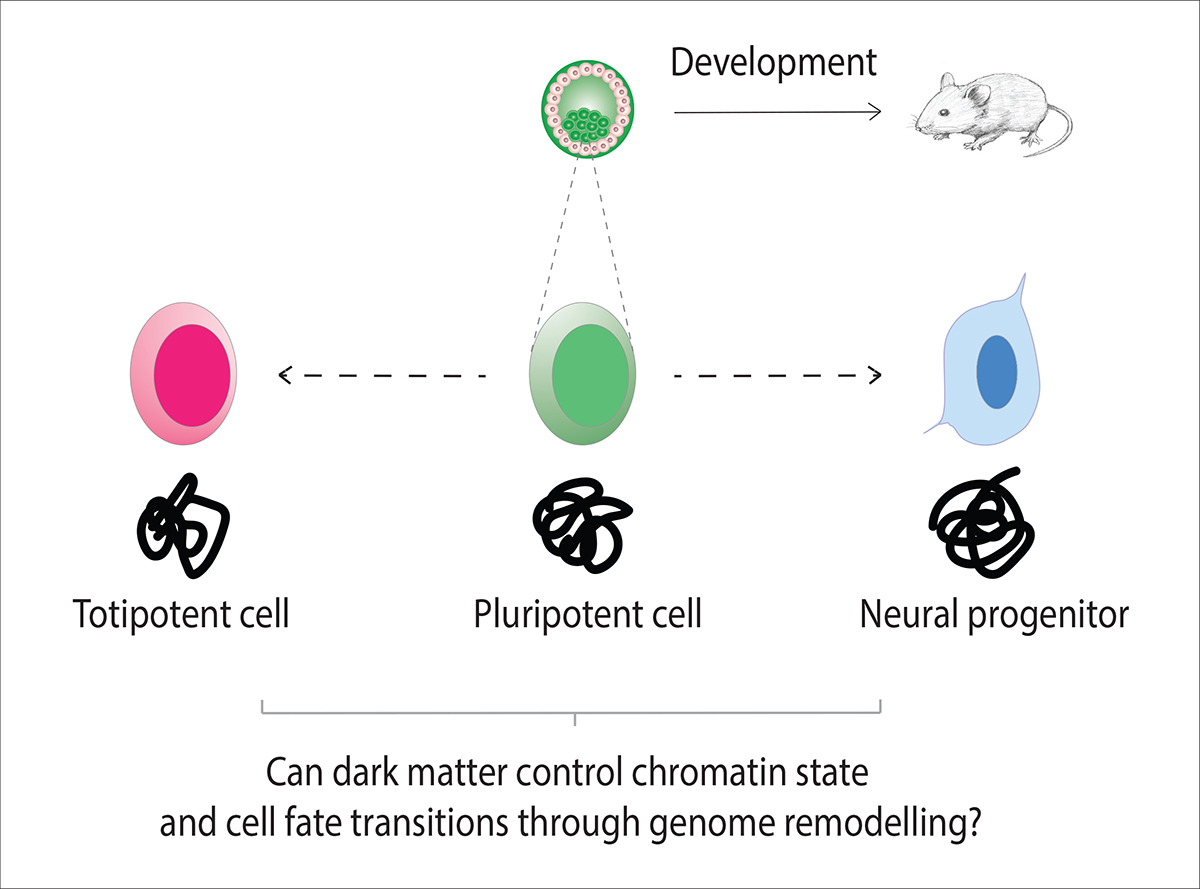
We are investigating how dark matter can control cell fate by using mouse embryonic stem cells to model developmental fate transitions in vitro. Mouse embryonic stem cells are derived from the inner cell mass of mouse embryos, at the blastocyst stage of development. The key feature of them is that they are ‘pluripotent’, which means that they can give rise to all cell types in an adult mouse. How are completely different cell types made when every cell contains the same genetic code or ‘blueprint’ to start with? It all depends on which genes are switched on and off to form a given cell type and this relates to the structure and function of the chromatin, which packages the DNA into chromosomes. In general, compacting and closing a segment of chromatin will switch off the genes that reside within it, whereas opening it will allow genes to become switched on.
Genome remodelling is whereby the global chromatin state of the cell changes to simultaneously switch on and off different sets of genes to control a cell fate transition. In the lab, we employ cell culture conditions that cause a pluripotent cell to differentiate into a neural progenitor cell, which provides us with a model to measure a cell fate transition. Neural progenitors are representative of cells committed to give rise to the mouse central nervous system.
Our work so far suggests that by manipulating dark matter, we can control whether or not cells will adopt a neural fate. We can also influence whether cells get ‘reprogrammed’ and adopt a cell fate resembling an earlier stage of mouse development – all through changes to dark matter. Understanding these processes will allow us to apply this knowledge to making more effective human stem cell therapies in the future. It will also enable us to decode dark matter and understand more broadly how gene networks are controlled.

TransposonsReprogram has received funding from the European Union’s HORIZON 2020 Research programme under the Grant Agreement no. 678350.
Please note: This is a commercial profile
© 2019. This work is licensed under CC-BY-NC-ND.