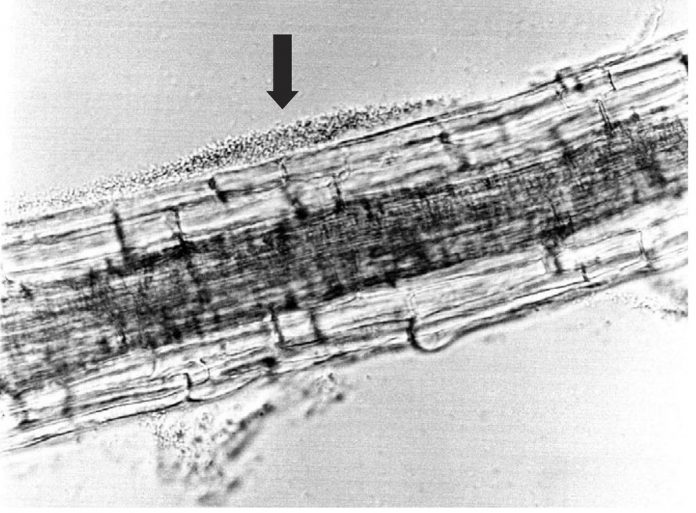
Ann G. Matthysse, Professor of Biology from The University of North Carolina at Chapel Hill provides an informative introduction to E. coli and diarrheal disease
Escherichia coli is a small gram-negative bacterium that lives in the gut of many mammals including humans. This bacterium has a fast growth rate. If given rich medium, it can double every 20 minutes. It is also easy to grow and needs no special nutritional supplements. Although it grows well at human body temperature (roughly 37°C), it can also grow at room temperature. It is a versatile bacterium and can utilise many substrates for growth. It can grow both aerobically and anaerobically (that is, it is a facultative anaerobe and can grow with or without oxygen). The bacteria grow well in liquid media with or without aeration. They also grow well on solid surfaces including laboratory agar medium made in Petri dishes. The laboratory strain of E. coli contains genes encoding about 4,000 proteins. The bacteria are flexible genetically and can exchange DNA via conjugation. In the laboratory, they can take up DNA from the environment if treated with divalent cations such as magnesium and calcium (transformation). There is evidence that they can also take up exogenous DNA in their natural habitat in the gut. There are a number of bacterial DNA viruses (bacteriophages) that can infect E. coli. These viruses can transfer DNA between bacteria. The genetic versatility and ease of working with E. coli have made it a model organism in the laboratory for more than 80 years. It has been used for many studies of bacterial physiology and genetics. Because it is so well understood, it is also a favourite organism to use as a vehicle for cloned DNA.
Strains of E. coli
There are a large number of strains of E. coli. Laboratory strains are somewhat atypical when compared to strains isolated from the gut of animals or humans or from the environment. The natural strains contain more genes than the laboratory strains. About 2,000 genes make up the core genome, which consists of those genes which any strain of E. coli must have to live and grow. The remainder of the genes vary between strains (there are more than 15,000 different genes which have been found in different strains)1. Genes found in only some strains include genes involved in pathogenesis and in survival in a mammalian host. This variable genetic make-up means that different strains of E. coli cause different diseases. Thus, there are diarrheagenic E. coli (DEC), uropathogenic E. coli (UPEC), and E. coli which cause meningitis. In this article, I will focus on the DEC strains.
Diarrheagenic E. coli (DEC) strains
DEC live in the gut of many animals, including cows, deer, and other grazing animals. Infection with strains that cause illness in humans may be asymptomatic in animals. The bacteria live in the gut and are excreted in the faeces. Meat in processing plants can become contaminated with DEC due to contamination of the meat by bacteria from the animal’s gut. In the natural environment, DEC excreted in the faeces need to survive and reach a new host. One survival strategy involves the bacteria binding to plant surfaces. When the plant is eaten by a potential host, the bacteria enter and grow in the host digestive tract. Humans are infected by DEC by eating contaminated meat or plants. In the last five years in the U.S., DEC has been transmitted by clover sprouts, chopped salad, Romaine lettuce, flour, ground beef and bison, soy nut butter, and alfalfa sprouts. The resulting disease outbreaks have resulted in recalls of the contaminated products whenever possible resulting in economic loss.
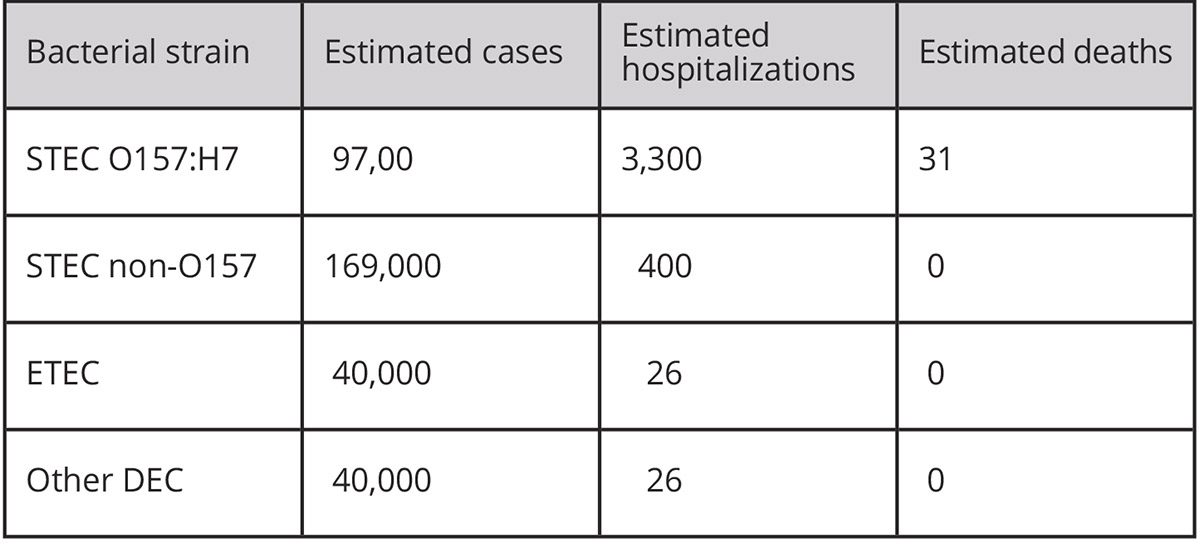
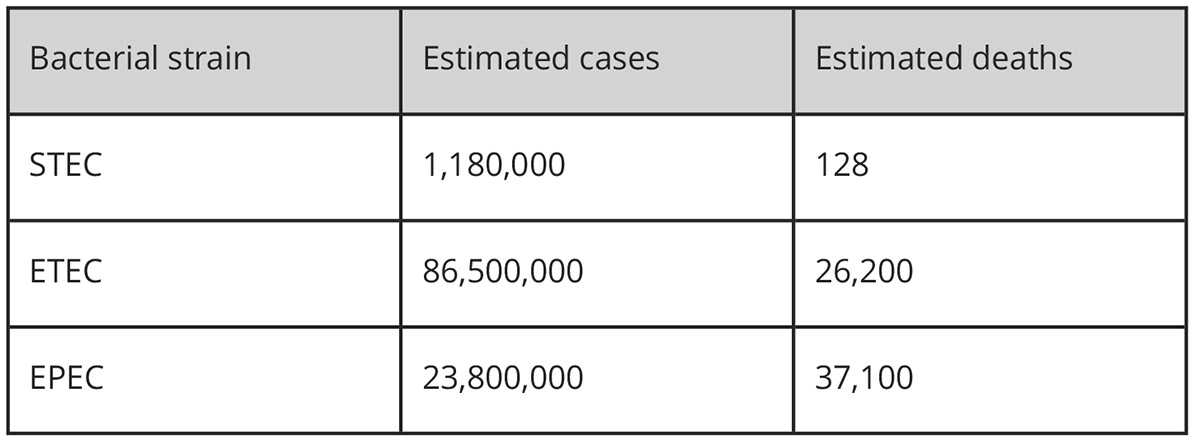
Disease caused by DEC is reported by the U.S. CDC (Center for Disease Control) in four categories: strains which carry the gene for Shiga toxin (STEC), one particularly virulent strain which carries the Shiga toxin gene and several other virulence genes (O157:H7), strains which carry genes for other toxins (ETEC), and other DEC strains. In developing countries with poor or no water purification and sanitation systems, DEC can easily spread from person to person via the faecal-oral route and cause large disease outbreaks. The estimated number of cases, hospitalisations and deaths caused by each of the groups of strains annually in the U.S. and worldwide is shown in the tables. As with all diarrheal disease, infections with DEC are most serious in babies and the elderly.
Looking ahead
This is the first of a series of articles. Future articles will consider transmission and reduction of DEC, and a survey of plant-borne human pathogens.
References
1 Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, et al. (2009) Organised Genome Dynamics in the Escherichia coli Species Results in Highly Diverse Adaptive Paths. PLoS Genet 5(1): e1000344. doi:10.1371/journal.pgen.1000344.
2 Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M, Roy SL, et al. Foodborne Illness Acquired in the United States—Major Pathogens. Emerg Infect Dis. 2011;17(1):7-15. https://dx.doi.org/10.3201/eid1701.p11101
3 WHO estimates of the global burden of foodborne diseases: foodborne disease burden epidemiology reference group 2007-2015. I. World Health Organization. ISBN 978 92 4 156516 5 © World Health Organization 2015 https://apps.who.int/iris/bitstream/handle/10665/199350/9789241565165_eng.pdf
Please note: This is a commercial profile