David S. Wishart discusses metabolomics, how the field is breaking into biochemical mysteries and the “dark metabolome”
The metabolome is defined as the complete collection of small molecule metabolites or chemicals that can be found in a cell, a tissue, an organism or even an environmental sample (such as soil or water). The study of the metabolome is called metabolomics. Metabolomics is a relatively new branch of the omics sciences that includes its better-known cousins: genomics, transcriptomics and proteomics. What makes metabolomics so different is that it focuses on small molecules (i.e., chemicals with a molecular weight less than 1500 Daltons) while the other omics fields focus on big molecules (i.e., DNA, RNA, proteins). What makes metabolomics so interesting is the fact that metabolites are the downstream products arising from the collective activities of the genome, the transcriptome and the proteome interacting with their environment. In other words, the metabolome is the closest thing we know of to a molecular phenotype.
In principle, if we could read out or “sequence” our entire metabolome, we should get a much clearer picture what is happening inside us than if we simply sequenced our genome. In this way, metabolomics could be used to precisely diagnose or predict diseases, to customize nutrition or accurately monitor our environment. The promise of metabolomics is huge. However, “sequencing” the metabolome is not as easy as sequencing DNA. Indeed, metabolomics requires the use of complicated mass spectrometers (MS) coupled to high-end liquid chromatography instruments. Even using the latest technologies and the best MS instruments available, scientists struggle to identify more than 500-600 compounds in any given biological sample. What’s more, most of the chemical signals that they see in their MS instruments are not identifiable. Indeed, it has been estimated that 90-95% of the MS peaks seen in most biological or environmental samples are not identifiable. They do not match to any of the known 100+ million chemicals on the planet!
What are these mystery compounds?
Biochemists and analytical chemists are stumped. They currently use the term “dark metabolome” to refer to the thousands of unknown, but clearly visible signals found in their MS instruments. The phrase “dark metabolome” grew from the term “dark matter’ that astronomers use to refer to the unknown, invisible matter that is thought to make up 85% of the universe. We know it’s there, but we just can’t figure out what it is. Just as dark matter gives physicists nightmares, so too does the dark metabolome haunt chemists.
How is it that despite 200 years of chemical research and the cataloguing of more than 100 million chemicals that we still can’t identify 95% of the compounds in ourselves, in our food or in our environment? What’s more, what are these unknown chemicals doing to us, to our fellow creatures and to our planet? Indeed, many of these unknown compounds consistently show up as being important biomarkers for disease, critical signals for biological processes, vital for sustaining health or persistently present in toxic extracts. Given its importance to human health and the planet’s health, the “dark metabolome” is something that we cannot ignore. So how do we illuminate the dark metabolome?
Traditionally, compound identification is a slow and arduous process. It often takes a highly skilled chemist up to 1-2 years to purify and positively identify a naturally occurring compound. Given that our MS instruments are telling us that there are literally millions of unknown, unidentified compounds found in our bodies, in our food and in our environment, the task of identifying the “dark metabolome” could take centuries of scientific effort and cost billions of dollars. Is there a better way?
How can computers help reveal the “dark metabolome”?
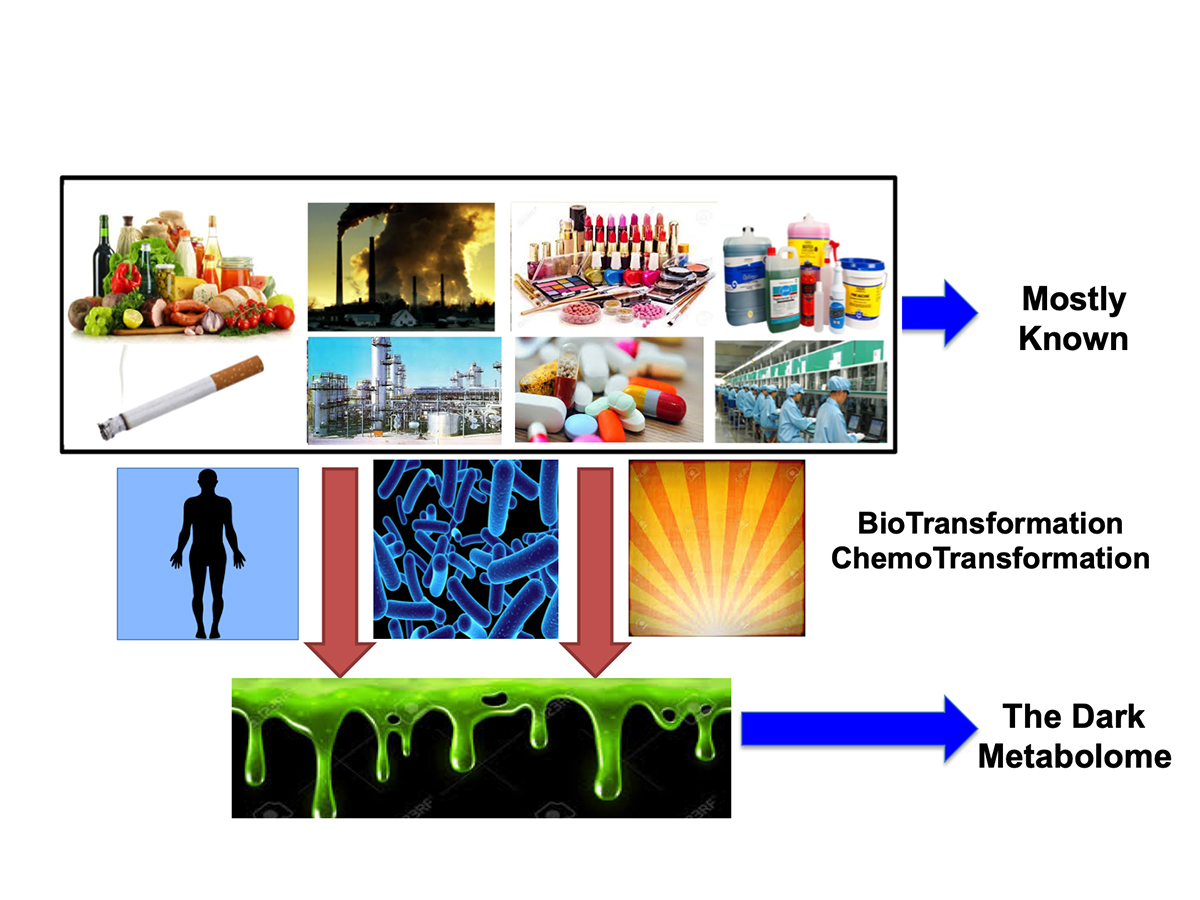
One approach that a handful of scientists, including ourselves, is to employ computers to help reveal the “dark metabolome”. This is sometimes called “in silico” metabolomics. It is potentially a far cheaper and a much faster approach to determining what these mysterious compounds might be. The concept is relatively simple. First it requires that we catalogue all of the known compounds coded by our genes (and the genes of our fellow creatures). These are called primary metabolites. Next it requires cataloguing all of the known compounds found in our food or added to our food. Third it requires gathering data on all of the compounds that are synthesized and released into the environment at measurable quantities (>1 tonne/year), such as herbicides, pesticides, dyes, drugs, cosmetics and paints.
Our lab has recently completed this process and we now have >500,000 “source” chemicals that should or could be found in our bodies, in our food or in the environment. This information is catalogued into publicly accessible databases called HMDB, FooDB, DrugBank, T3DB and ContaminantDB. The next step involves using computers to predict the reactions that these source chemicals go through as they are metabolized by our bodies or by the environment. This is called “in silico” biotransformation or metabolic fate prediction. My lab has recently developed an open-source software tool called BioTransformer to accurately perform this process. It uses machine learning, pattern recognition techniques and specially hand-crafted rules learned from thousands of known biotransformation reactions to predict how compounds are metabolized in the stomach, liver and gut as well as how they are metabolized in the soil and water.
Using BioTransformer we have taken the 500,000 source compounds and succeeded in predicting the structure of >2 million new compounds that are biological feasible but are not in any known chemical database. A second iteration of BioTransformer, to be finished later this year, is expected to generate >10 million new, biologically feasible compounds. Based on some early results we expect that >50% of the “dark metabolome” corresponds to these computer-generated compounds.
Our task, and that of our fellow chemists, is to confirm that these hypothetical compounds exist using various computer-aided spectral prediction and spectral matching techniques. Once these are identified, the real adventure can begin as we can truly start to “sequence the metabolome” and figure out exactly what these compounds do and why the “dark metabolome” seems to play such an important role in human health and disease.
Please note: This is a commercial profile