Dr Kristen Kroll from Washington University School of Medicine discusses new approaches for studying autism and other neurodevelopmental disorders by using human cellular models of brain development
Autism spectrum disorder (ASD) is among the most prevalent neurodevelopmental disorders (NDDs), with an estimate of 1 in 54 children in the United States. It can co-occur with other NDDs, including intellectual disability, attention deficit hyperactivity disorder, and developmental delay, and represents a significant burden for affected individuals, their families, and society.
NDDs have been diagnosed in individuals of all countries, ethnicities, and socioeconomic backgrounds, and represent a significant global health challenge.
The complex biology of NDDs
Despite their widespread prevalence, the causes of NDDs are complex and remain poorly understood, often disrupting multiple aspects of brain development. Both clinical and genetic heterogeneity make it challenging to understand the basis of these disorders. For example, individuals with ASD have a wide range of symptoms and phenotypes, ranging from mild social deficits to severe cognitive impairment and disability.
ASD is also genetically complex, with sequencing of human subjects implicating hundreds of genes in risk. Adding to this complexity, ASD risk is often polygenic, involving mutations in multiple genes, or mutations in the non-protein coding portion of the genome. Such non-protein coding mutations may contribute to risk by altering regulation of gene expression.
How can human cellular models of altered brain development be used to understand autism and other neurodevelopmental disorders?
The clinical and genetic complexity of NDDs poses challenges for studying these disorders. Many risk-associated mutations are also human-specific, such that these disorders often cannot be modelled in animals; however, lack of access to the developing human brain precludes direct study. Given these challenges, human cellular models play a central role in NDD research. These models are derived from human subjects with NDDs, by converting a patient’s own skin or blood cells into induced pluripotent stem cells (iPSCs) used to model aspects of brain development. These human models faithfully recapitulate the complex human genetics that underlie NDDs.
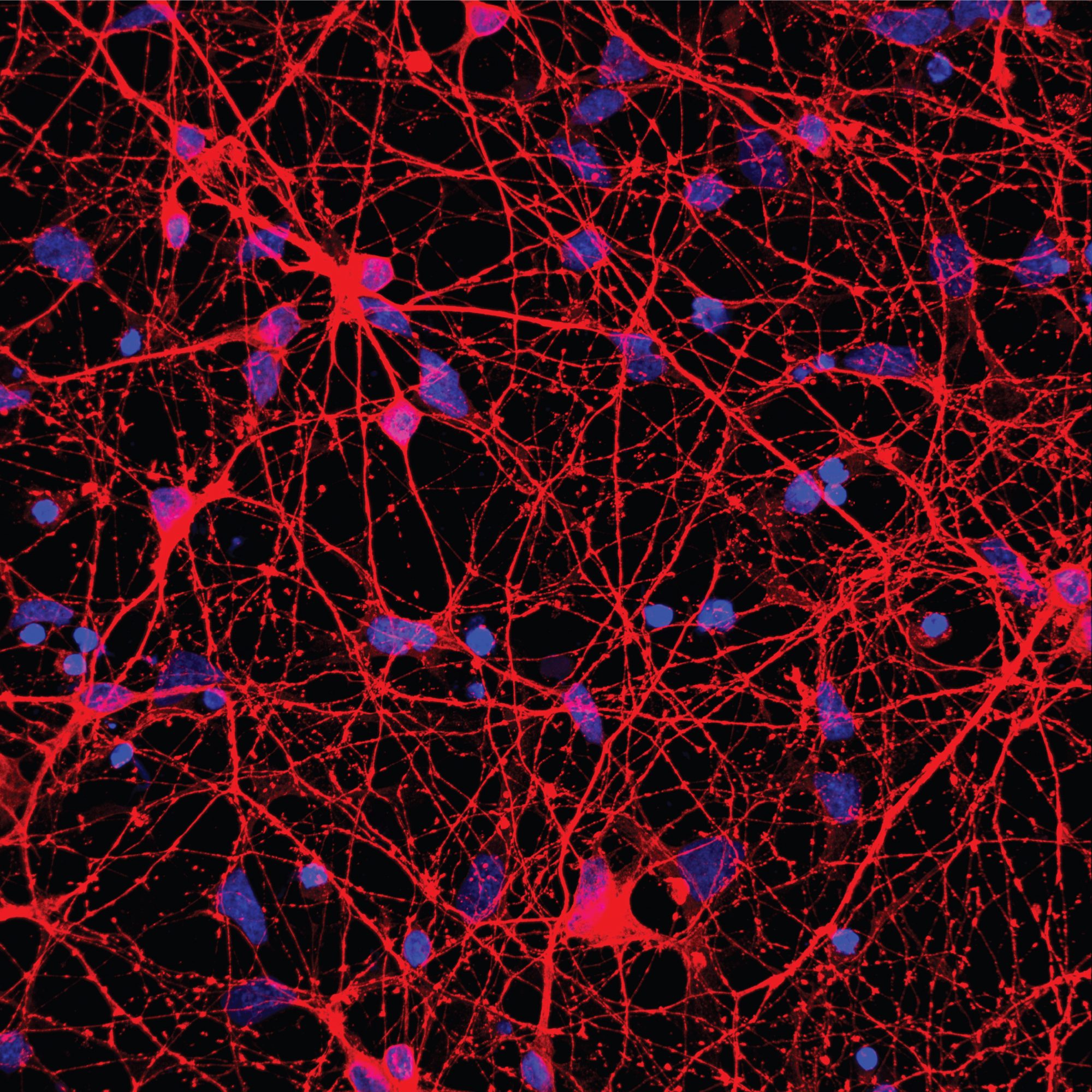
Neurons generated from human stem cells (Image: © Gareth Chapman)
In ongoing studies, supported by the National Institute of Neurological Disorders & Stroke (NINDS) and National Institute of Mental Health (NIMH) (parts of the National Institutes of Health in the U.S.), we are using human cellular models to define mechanisms of brain development and determine how their disruption causes NDDs. For example, we characterised gene regulatory networks that control the development of cortical interneurons. These inhibitory neurons are frequently disrupted to cause NDDs, but the developmental programs that generate them are complex and remain largely uncharacterised. We found that CHD2, a protein mutated to cause NDDs, works with the transcription factor NKX2-1 to regulate gene expression during interneuron development.
We are currently studying many aspects of this developmental programme and exploring how its disruption contributes to NDDs.
New cellular models of autism spectrum disorder
Human cellular models can be used to study many aspects of NDDs. For example, while autism risk is highly heritable and usually involves multiple genetic risk factors, most current research focuses on de novo (new rather than inherited) cases involving rare single gene mutations. Therefore, we recently generated iPSC models of multiplex autism, the most common form of ASD involving inherited, polygenic risk.
We derived models from several family members with different degrees of inherited ASD (severe, intermediate, unaffected) and differentiated these into cortical neurons. By comparison with models from both unaffected relatives and unrelated, unaffected individuals, cellular models from ASD-affected family members exhibited increased cell death. Models from ASD-affected individuals also had mis-regulated expression of genes associated with ASD, neural development, behaviour, and cognition. This work demonstrates that, even for genetically complex, heritable forms of ASD, we can identify cellular and molecular abnormalities associated with diagnosis.
Cellular models can also be used to understand why neuropsychiatric disorders arise with variable penetrance (e.g. the extent to which genetic risk causes a disorder). For example, copy number variants (CNVs) that micro-duplicate a portion of the genome containing the CHRNA7 neuronal channel gene cause severe phenotypes in some individuals but not in others.
To define the basis of this phenomenon, we recently derived cellular models from several family members carrying the same duplication of CHRNA7, with some having an ASD diagnosis, while others are clinically unaffected. Models with CHRNA7 duplication had elevated CHRNA7 gene expression, regardless of patient diagnosis. However, models from an ASD-affected individual had many phenotypes not seen in models from an unaffected family member with CHRNA7 duplication or unrelated individuals without CHRNA7 duplication. These included increased neuronal progenitor cell proliferation but defective neuronal differentiation, maturation, and migration, and increased cell stress. Gene expression related to these processes was also reduced in this model. However, individuals with a CHRNA7 duplication but no clinical signs of ASD may have compensatory mechanisms that enable normal brain development. Indeed, we found the model from an unaffected individual with CHRNA7 duplication instead had elevated expression of genes in the same neuronal differentiation and maturation pathways. Finally, both models with CHRNA7 duplication, with or without an ASD diagnosis, had altered neuronal function consistent with elevated CHRNA7 channel activity.
This work demonstrates that human cellular models can define phenotypes associated with variable phenotypic penetrance among individuals with the same CNV. Some diagnosis-associated anomalies could be rescued with drugs targeting the affected pathways (e.g. elevated cellular stress was suppressed by the drug JTV519). Therefore, these findings suggest avenues for intervention, both for carriers of this duplication and other CNVs that cause related disorders.
Future prospects
Human cellular models provide an essential research tool for studying NDDs. These renewable, sharable models can reveal contributors to disease. Building and comparing many different types of NDD models can uncover commonly disrupted aspects of brain development. These models can then be used to screen for drugs or molecular therapies to reverse diagnosis-associated phenotypes.
Together, these studies may suggest ways to target and correct disrupted aspects of brain development and function that contribute to NDDs, presenting avenues for developing interventions for affected individuals.
Please note: This is a commercial profile
© 2019. This work is licensed under a CC BY 4.0 license.