Regina Fluhrer from the LMU & DZNE in Munich explains an aspect of health research that concerns how proteases embedded in cellular membranes hydrolyse other proteins and the challenges they must overcome
The concept of proteolysis was first discovered in 1836, when Theodor Schwann, a German researcher, purified pepsin as an enzyme capable of fragmenting nutrient proteins in the intestine. Peptide bonds, the chemical bonds that connect amino acids within a protein, are very stable and hydrolysis, the breakage with water, is not easy. This is important because it allows, for instance, that we can shower in the morning without our skin dissolving. However, it is a hurdle if it comes to degradation and, thus, cleavage of these bonds, since energy, the so-called activation energy, needs to be invested. Here, proteases come into play: with their special active site, also known as the catalytic centre, they interact with their substrates forcing them into a special confirmation that reduces the activation energy allowing hydrolysis of peptide bonds within milliseconds that without this catalysis would take hundreds of years and life wouldn’t be possible.
To date, more than 500 human proteases, with specificity for an even greater number of substrates are known. However, the vast majority of these proteases are soluble proteins that are active in an aqueous environment within cells and tissues or in body fluids like blood.
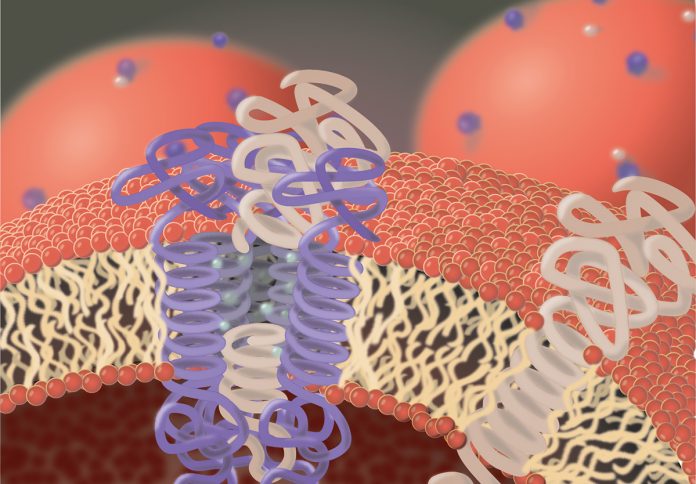
In 1997, more than 150 years after the discovery of the first protease, it became evident that proteolysis also occurs within cellular membranes. Membranes of cells are composed of lipids that arrange as bilayer to separate the inner of a cell from its surroundings, but also allowing a limited and very selective permeability, particularly for small molecules. To allow passage of larger or charged molecules, like glucose and certain ions and to enable cells to communicate with their environment, various proteins are integrated into cellular membranes.
Many of these proteins span the membrane with domains that are mainly composed of amino acids that allow interaction with lipids. These proteins are termed integral membrane proteins. Like all cellular proteins, integral membrane proteins are subject to a permanent turnover that requires de novo synthesis and subsequent degradation. Protein degradation requires proteases since they are the only players in a cell capable of hydrolysing peptide bonds.
Proteases capable of cleaving peptide bonds in membrane-embedded protein domains, face specific difficulties compared to proteases cleaving soluble proteins: i) the peptide bonds in their substrates are embedded in lipids and thus access of the protease to its substrate is much more difficult than in solution. ii) lipid-embedded protein domains tend to form ordered helical structures, which again aggravates cleavage. iii) water, critical for hydrolysis, is much less abundant in membranes. iv) diffusion rates in lipid environment differ from that in solution.
Remarkably, despite these additional challenges, intramembrane proteases apply similar catalytic strategies as their soluble cousins. Based on the amino acids or co-factors critical for catalysis in their active centre four major classes of intramembrane proteases have been identified: metallo, serine, glutamyl and aspartyl intramembrane proteases.
The peculiarity of these proteases is that their active site, like the cleavage site in their substrates, localises to domains embedded in the cellular membrane. In addition, intramembrane proteases comprise further membrane-spanning domains that allow the formation of a pore-like structure in the lipid environment. Thus, intramembrane proteases combine protease features with that of transporters and channels as they are, for instance, used to carry glucose or sodium ions over membranes. Their membrane-spanning domains allow contact with the integral membrane domains of their substrates and most likely by a lateral opening of the pore-like structure the substrate gains access to the protease active site. This structural layout also allows access of water molecules so that proteolysis can take place. Although the mechanism of the actual peptide bond hydrolysis is similar to that observed for soluble proteases, kinetics of substrate recognition and cleavage by intramembrane proteases seems to be fundamentally different and are far from being completely understood.
Cleavage of integral membrane proteins by intramembrane proteases liberates protein fragments on both sides of the membrane. Thus, intramembrane proteases not only initiate degradation of membrane proteins but also produce protein fragments that are capable of transmitting signals across membranes. In that context, S2P, a metallo intramembrane protease, has been linked to the regulation of cholesterol synthesis, SPPL3, an aspartyl intramembrane protease to the regulation of protein glycosylation and various intramembrane proteases contribute to cell differentiation (read more in the January 2019 edition of Open Access Government).
Moreover, intramembrane proteases contribute to the proper function of the immune system (read more in the April 2019 edition of Open Access Government) and are implicated in the development of neurodegenerative diseases (read more in the July 2019 edition of Open Access Government), cancer and infectious diseases (read more in the October 2019 edition of Open Access Government).
Compared to soluble proteases, which have become important and valuable targets for treatment and prevention of human diseases, like HIV infections, high blood pressure or stroke, intramembrane proteases are only known for a short period of time. To utilise intramembrane proteases in the future as targets to treat or even cure diseases, it is indispensable to understand their cleavage mechanisms, substrate repertoires and physiological function in depth. Therefore, much more basic research on these fascinating enzymes will be required to raise this treasure.
Further Reading
Paschkowsky S, Hsiao JM, Young JC, Munter LM. The Discovery of Proteases and Intramembrane Proteolysis. Biochem Cell Biol. 2018 Aug 13.
Mentrup T, Fluhrer R, Schröder B. 2017. Latest emerging functions of SPP/SPPL intramembrane proteases. Eur J Cell Biol. 96(5):372-382.
Please note: this is a commercial profile
Prof Dr Regina Fluhrer 
Biomedical Center (BMC), Ludwig Maximilians University of Munich (LMU) & German Center for Neurodegenerative Diseases (DZNE)
Tel: +49 89 440 046 505
regina.fluhrer@med.uni-muenchen.de
https://www.biochemie.abi.med.uni-muenchen.de/fluhrer_lab/index.html
Intramembrane Proteolysis illustrated by Charlotte Spitz