Professor Wolfgang Streit from the University of Hamburg details how functional metagenomics applies to modern biotechnology, new drugs and much more
Life on earth is dominated by prokaryotic microorganisms (bacteria and archaea) and it is estimated that more than 1030 microbial cells are living on our planet1. They harbour a sheer unlimited number of genes encoding a tremendous and unseen metabolic and catabolic potential. The richness and diversity in the genes of all microbes are by far much higher than those of all plants, animals or human genomes together. A recent study estimated the global soil metagenome to harbour at least 160 million soil specific genes2 and for the marine system, a comparable number of roughly 111 million specific genes was estimated3. Because of the incredibly huge prokaryotic biomass, their global importance for all major bio-geochemical cycles, their impact on production of climate-relevant gasses such methane or CO2, their well-known roles in infection, pathogenicity, symbiotic interactions and their occurrence in all microbiomes there is a very vigorous and permanent interest in understanding the functions of genes and individual microbes in their habitats.
Furthermore and because of their truly high metabolic diversity microorganisms are a very attractive and promising resource for the identification of novel biocatalysts, urgently needed antibiotics, anti-cancer drugs and other valuable biomolecules. In fact, microorganisms code for the blueprints of many innovative products and novel sustainable processes in their genomes and they offer solutions to some of the most pressing environmental and societal problems we are currently facing (e.g. plastic pollution, lack of environmentally friendly pesticides, and no new antibiotics in sight). In other words, marine and terrestrial microbial resources offer the keys and answers to future biotechnology processes and societal questions.
It is, however, not easy to unleash this vast potential and access these microbes. Mainly two reasons hinder us currently from using this immense potential and deciphering microbial genes and functions on a global scale. First, only less than 1% of all microbes can be cultivated with traditional but also highly sophisticated methods. Thus, the vast majority (>99%) of all microbes remains untouched with respect to their functional roles in ecology and/or their use for biotechnology or any other application. Second, we have a large gap in knowledge on the functions of genes. A large fraction (>50%) of all genes encoded in the global metagenomes and genomes of individual microbes is unknown with respect to their function and even more importantly, many genes have not yet been identified. We call this large fraction of unknown genes ‘dark matter’ and we are only slowly beginning to develop methods to address these problems
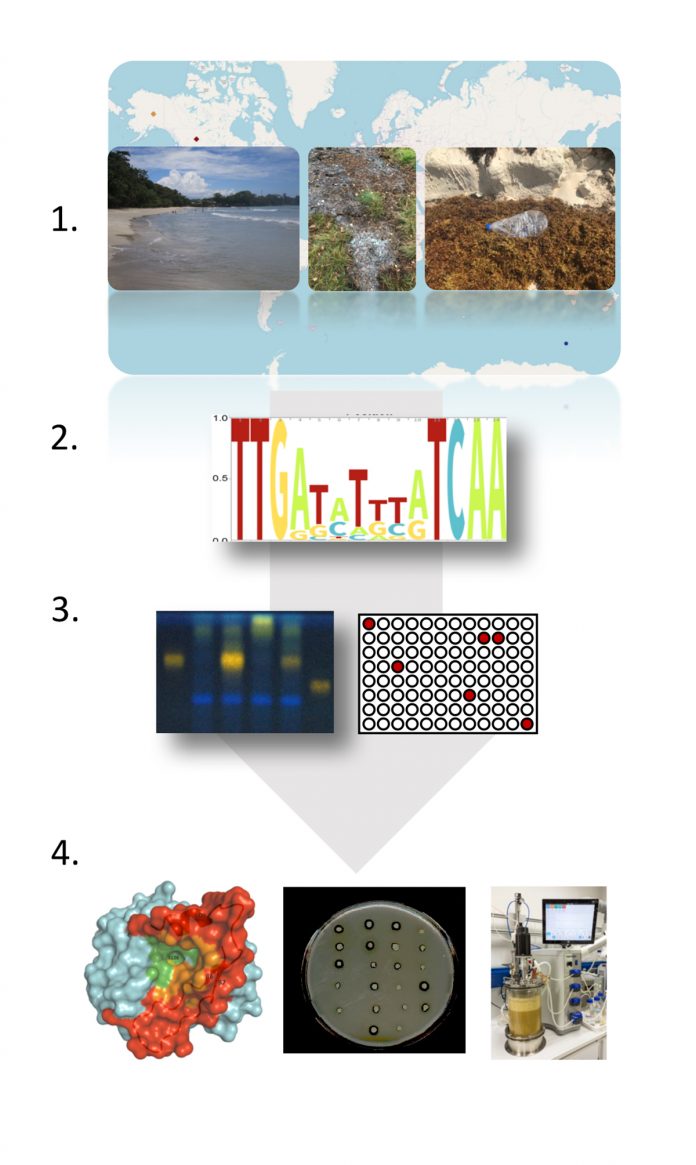
Functional metagenomics is the key technology that offers solutions to these challenges. First, it offers a solution for the poor cultivation rates and second, while permitting access to the non-cultivated microbial diversity, it assigns functions to hitherto unknown genes, enzymes and metabolic pathways and it allows identifying novel protein families with novel functions. Functional metagenomics develops and uses methods to exploit microbial communities or single microorganisms without the need to cultivate them. Thereby, this groundbreaking technology exploits the DNAs, RNAs, the proteomes and metabolomes of the globally distributed microorganisms. This is mostly done by extracting samples directly from nature and then analysing these using an array of modern ‘omics’ and analytical tools and technologies including next-generation sequencing, metabolomics, proteomics, bioinformatic approaches, structural biology and others. During the onset of this research, this was achieved by focusing on a single sample or just a few samples obtained from a confined source. Thanks to the setup of global databases today, this can be accomplished on a global level by accessing the respective databases such as NCBI, IMG or MGRAST data repositories. Thereby, very often, novel globally important organisms, new genes coding for novel enzyme families, complete pathways for antimicrobials or other drugs are identified. Notably, during this process very often, functions are assigned to genes being part of the dark matter.
Functional metagenomics is a very modern key technology enabling the development of novel biotechnological and pharmaceutical products. Since the term ‘metagenome’ was coined more than two decades ago, it has resulted in the rapid identification of many thousands of novel biomolecules and biocatalysts with potential value to many bioindustries for the development of more sustainable products. This includes robust and versatile enzymes for food, feed, and textile industries, paper production, fine and bulk chemicals and cosmetic production. By delivering such a wealth of novel biocatalysts, the technology has led to a much better understanding of the evolution, the structure and the function of biocatalysts and this knowledge have advanced our efforts to identify novel biocatalysts significantly. Thereby, it has further helped us to rapidly integrate enzymes in the novel production process.
Functional metagenomics has given us the first clues to pressing societal problems. For example, the removal of plastics from the environment is an urgent problem that if not solved will have a severe impact on our food and feed chain and our well-being. Using functional metagenomics, we have recently identified hundreds of novel enzymes involved in PET degradation4. This is a major breakthrough using functional metagenomics and with respect to environmental pollution.
The technology has also given us access to novel antibiotics or other drug molecules with new and or improved traits. It strongly advances the speed of natural product discovery and at the same time, it allows the identification of completely novel structures. For instance, recently Iqbal and colleagues identified a novel antimicrobial compound designated metatricycloene5. This compound was identified through the mining of over 1.2 Mio clones and it represents the first example of a new class of antibiotics identified using functional metagenomics.
Besides the above-listed examples, functional metagenomics has further provided recent discoveries on the physiology of non-cultured microbes and their significance for global biogeochemical processes and it has significantly increased our knowledge on host-microbiota interactions, infection and pathogenicity.
Thus, in summary, functional metagenomics is a very modern key technology driving the discovery of knowledge for basic research and advancing the identification of novel protein families, biocatalysts and other valuable biomolecules. Thereby, providing access to innovative bioprocesses leading to the development of novel drug molecules and helps to provide a solution to some of the environmental and societal needs.
References:
1 Whitmannn et al. PNAS, 1998, 95 (12) 6578. 6583; https://doi.org/10.1073/pnas.95.12.6578
2 M. Bahram, et al., 2018;560(7717):233-237. DOI: 10.1038/s41586-018-0386-6.
3 Sunagawa et al., Science, 2015:348, 6237, 1261359 DOI: 10.1126/science.1261359.
4 Danso et al., Appl Environm Microbiol 218, 84(8), DOI: 10.1128/AEM.02773-17.
5 Iqbal et al., J. Am. Chem. Soc., 2016, 138 (30), pp 9341–9344 DOI: 10.1021/jacs.6b02921.
Please note: this is a commercial profile
Professor Wolfgang Streit
Department of Microbiology and Biotechnology,
University of Hamburg
Ohnhorststrasse. 18, 22609 Hamburg
Tel: +49 40 42816 463