Dr Kim Hammond-Kosack at Rothamsted Research highlights an aspect of plant pathology that concerns the importance of finding new ways to disarm old enemies in wheat diseases
Wheat is the number one arable crop grown in the UK and Europe, and globally, the wheat crop provides 20% of the total calories consumed by humans each day. Wheat is also a valuable source of animal feed and is increasingly used in a wide range of manufacturing processes. The EU harvested 142.6 million tonnes of wheat in 2017.
Currently, wheat yields are not resilient to adverse weather conditions or various biotic and abiotic stresses.
As a result, ‘on farm’ yields remain static. Each season because of diseases caused by microscopic pathogens, predominantly specialist infectious fungal, bacterial and virus species, potential wheat yields are reduced by 16 to 25%. Pathogens rapidly grow and reproduce within wheat plants and spread disease to neighbouring fields and regions thereby causing disease epidemics and pandemics if left unchecked. Collectively, these pathogens reduce crop canopy, root system and flower health thereby lowering grain yield, quality and sometime grain safety.
Within the wheat pathogenomics research team at Rothamsted we focus on several pathogenic fungi of global importance. In this article we will feature two persistent disease problems of fungal origin which are causing the agrochemical industry, plant breeders, agronomists and farmers frequent headaches. The first leaf blotch disease is caused by Zymoseptoria tritici and the second Head blight disease is caused by a group of Fusarium species (Figure 1).
Our multi-disciplinary team includes plant pathologists, molecular biologists, geneticists, computational biologists and field crop scientists. We are pursuing a two-pronged experimental approach. On the one hand we investigate how fungi attack wheat and cause disease by discovering the genes, proteins and chemical metabolites involved, as well as defining the underlying interconnecting networks which co-ordinate the early infection process.
In concert with this, we are exploring the molecular processes used by wheat cells to perceive fungal pathogen attacks as well as dissecting the plant defence components that are activated to rapidly stop fungal growth and minimise disease formation. In recent years, our discovery research toolbox has been substantially improved thanks to the full sequences of pathogen and wheat genomes becoming available (i.e. their DNA blueprints).
Finding pathogen genes required for stealth or attack
To be successful, a pathogen needs initially to attach itself to, and then grow along, across and ultimately into, plant tissues. Whilst doing so it must also mask its presence and remain symptomless, until its size and strength becomes sufficient to overwhelm the plant and cause disease. Our group has identified several key genes from both Fusarium and Zymoseptoria species which are important for these events. By randomly inducing mutations into the genome of Zymoseptoria and then testing for mutants which could not grow on leaf surfaces, we’ve identified a key virulence gene GT2 (a specialist enzyme called glycosyltransferase 2).
The identical Fusarium gene is also crucial for the infection of wheat floral tissues. Inactivation of these genes would likely protect plants from the corresponding diseases. We’ve also identified a key “stealth” gene called 3LysM which prevents Zymoseptoria being recognised early on by the plant’s defences, thus giving it time to build up inside the infected wheat leaves. Blocking this fungal gene would also likely prevent disease.
Finding wheat genes required to stop pathogens
To be successful, a plant needs to recognise an invading pathogen and rapidly switch on an effective defence response as well as prepare for subsequent attacks.
Despite Zymoseptoria being such an important wheat pathogen, only a small number of genes for resistance to Zymoseptoria have been identified so far and surprisingly very little is known about how these genes actually operate to protect wheat.
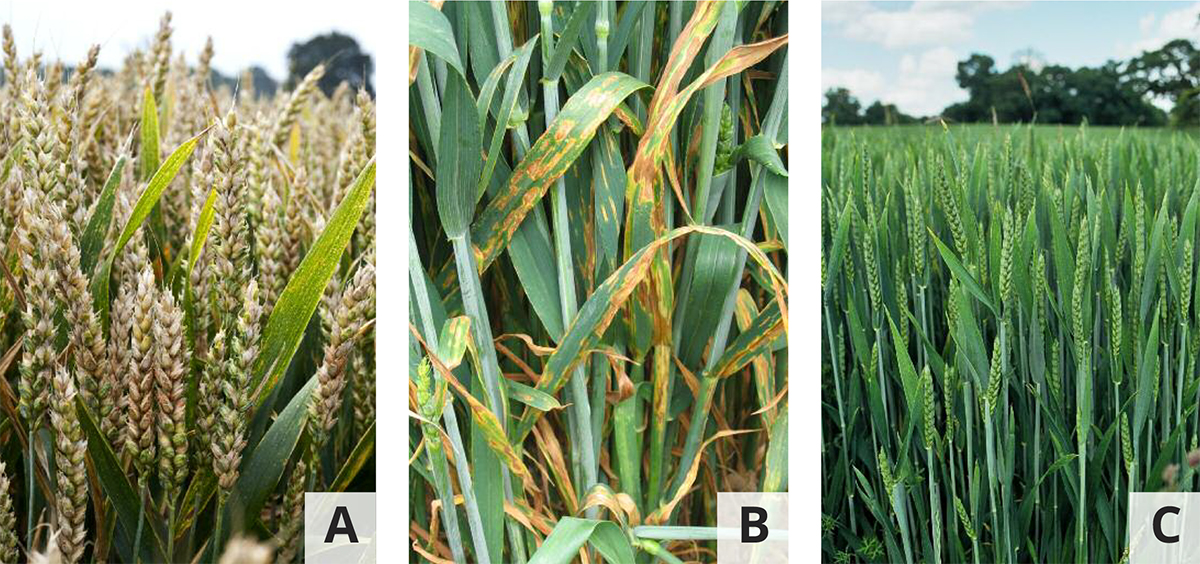
To help us answer some of these questions, we isolated the very first Zymoseptoria resistance gene from wheat known as Stb6. This gene belongs to a large previously uncharcterised gene family in wheat, codes for a cell wall residing immune receptor protein that senses a protein secreted by Zymoseptoria during attempted infection and triggers defence.
Unravelling the molecular mechanism of Stb6 mediated resistance will help inform future effective strategies for disease control. Moreover, the knowledge already gained in our studies will help to isolate additional resistance genes for deployment through breeding in wheat improvement programmes.
Outlook
The most sustainable solution to reduce plant diseases is through delivering life-long protection within or on the planted seed. This can be achieved by combining a triad of approaches. Through plant breeding by ‘stacking’ or ‘pyramiding’ multiple disease resistance genes that activate different plant defences against the same pathogenic species.
By removing or inactivating disease susceptibility genes that were unwittingly incorporated into elite cultivars by breeders when introducing another, beneficial trait. Through the application of novel chemicals or biological agents to the seed in coatings that go on to combat infections throughout the life-span of each plant by reducing the effectiveness of pathogen stealth or attack genes.
References
https://onlinelibrary.wiley.com/doi/full/10.1111/j.1364-3703.2011.00783.x
http://www.plantphysiol.org/content/early/2011/04/05/pp.111.176347?versioned=true
https://apsjournals.apsnet.org/doi/10.1094/MPMI-07-13-0201-R
https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1006672
https://www.nature.com/articles/s41588-018-0051-x?
https://science.sciencemag.org/content/361/6403/eaar7191
https://designingfuturewheat.org.uk/
http://www.wgin.org.uk/
Dr Kim Hammond-Kosack
Research Programme Leader Wheat Pathogenomics
Rothamsted Research
Tel: +44(0) 158 293 8240
kim.hammond-kosack@rothamsted.ac.uk
https://www.rothamsted.ac.uk/our-people/kim-hammond-kosack
*Please note: This is a commercial profile