Professor Yoshiaki Hasegawa, Aichi Gakuin University, Department of Microbiology, School of Dentistry, discusses the structure and biogenesis mechanism of Mfa1 fimbriae from the periodontal pathogen Porphyromonas gingivalis
Periodontal disease is an inflammatory disease caused by infection of the gums and results in the collapse of the structures supporting teeth. It is very common worldwide, affecting as much as 50% of the global population.
Moreover, the disease has been shown to be associated with various systemic diseases, including, cardiovascular diseases, diabetes, preterm birth, pulmonary disease, and rheumatoid arthritis. Periodontitis is a chronic inflammatory lesion-induced and maintained by the polymicrobial biofilm formed on teeth, according to the polymicrobial synergy and dysbiosis hypothesis.
In recent years, microbiome analysis and animal models of periodontal disease have led researchers to propose the hypothesis that Porphyromonas gingivalis is a keystone pathogen in causing dysbiosis of the dental plaque bacterial flora.
The periodontal pathogen Porphyromonas gingivalis
- gingivalis, a Gram-negative anaerobic bacterium, produces several known potential virulence factors: Fimbriae, proteases like gingipains, and lipopolysaccharides. Fimbriae are fibrillar structures formed by the polymerization of fimbrilins on the bacterial surface. They play a pivotal role in P. gingivalis colonization, invasion, establishment, persistence within the host, and evasion from immune system destruction. P. gingivalis typically expresses two different types of fimbriae, FimA and Mfa1. FimA and Mfa1 fimbriae are primarily composed of polymers of the FimA and Mfa1 proteins encoded by the fimA gene and the mfa1 gene, respectively.
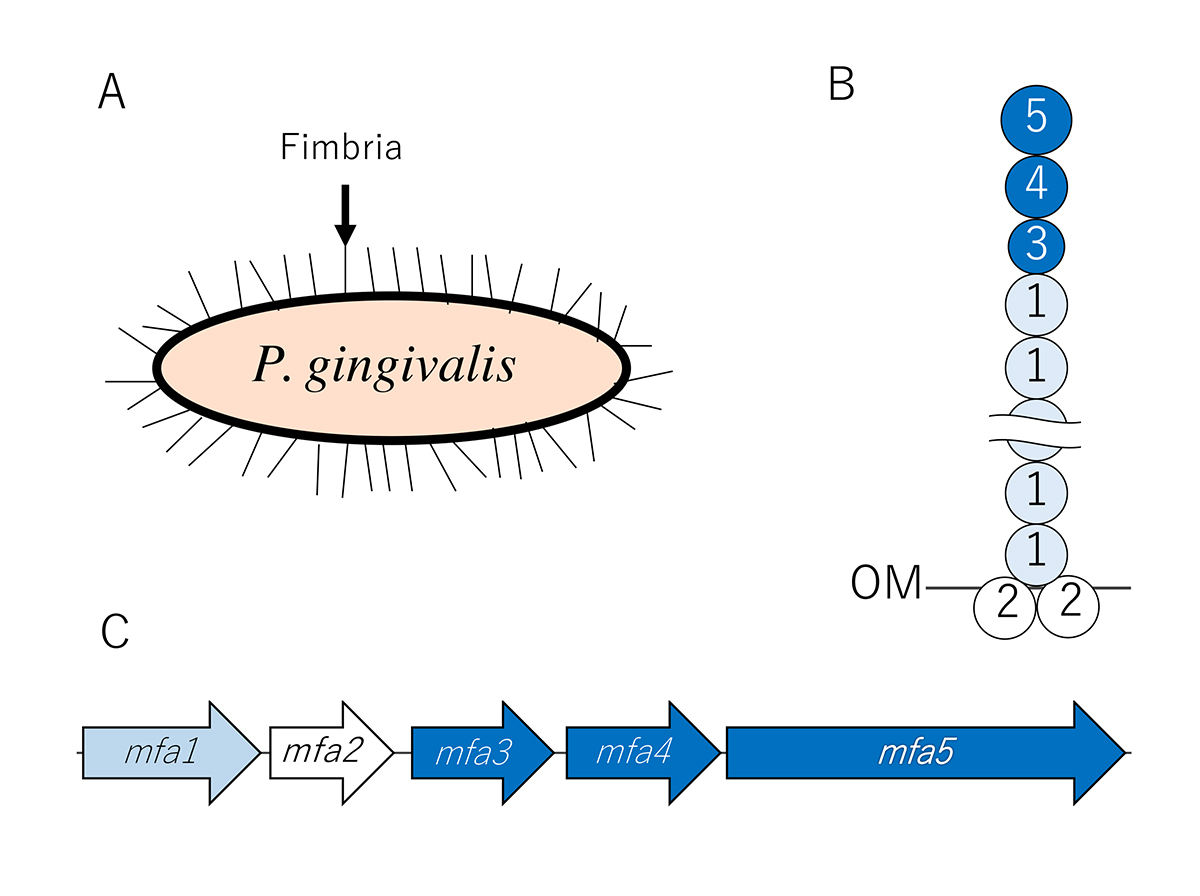
In addition to structural proteins, mature FimA and Mfa1 fimbriae contain the accessory proteins, FimC-E and Mfa3-5, respectively. However, the structure and biogenesis mechanisms of both fimbriae have not been closely examined.
Structure and biogenesis mechanism of Mfa1 fimbriae
Since 2007, we have been analysing the structure and biogenesis mechanism of Mfa1 fimbriae. We have determined that the mature Mfa1 fimbria is generated via an mfa gene cluster comprised of five proteins: Mfa1, Mfa2, Mfa3, Mfa4, and Mfa5. To examine the roles of Mfa2-Mfa5, the deficient mutant strains of these genes were constructed. In our results, we found that the main shaft portion consists of Mfa1, while Mfa2 is located in the basal portion of the fimbriae and functions as an anchor and an assembly and elongation terminator.
In addition, Mfa3 is localised in the distal tip portion of the Mfa1 fimbriae, and thus may play a role as a ligand to receptors on host cells and other oral bacteria. Mfa3, Mfa4, and Mfa5 all participate in the assembly of an accessory protein complex on the tips of fimbriae. Mfa5, which contains a C-terminal domain and von Willebrand factor type A domain, is translocated to the cell surface by the Type 9 secretion system. Our proposed a structure model of Mfa1 fimbria is shown in Figure 1.
Benefits of our project
Since the protein located at the tip of the fimbria often functions as an actual adhesion in fimbriae of many pathogenic bacteria, it is thought that the tip proteins of Mfa1 fimbria of P. gingivalis play an important role in the colonization of periodontal tissue. If we are able to identify the binding partner of the tip accessory proteins of Mfa3-5, we will be able to reveal the colonization mechanism of this bacterium.
Furthermore, this information would allow us to devise a new therapeutic strategy for the prevention of colonization. In the future, we hope to lead in the development of locally administered treatments, such as drugs or peptides that can inhibit the function of P. gingivalis fimbria. These treatments could be applied after professional mechanical tooth cleaning, such as scaling by a dentist. We hope that the results of these research projects will lead to the development of new therapeutic and preventive drugs for periodontal disease targeting Mfa1 fimbriae of P. gingivalis.
Recently, Xu et al. (2016) clarified the crystal structure and polymerisation mechanism of a group of proteins which predominantly exist in human microbiomes and are presumed to be a fimbrilin of class Bacterioidia (of which P. gingivalis is also a member). In addition, the fimbrae of this family did not belong to any current type of bacterial fimbriae (type I to IV) and were, therefore, classified as V-type fimbriae. We hope that by exploring these research avenues, future research will be able to lead the development of a new treatment strategy that controls the bacterial flora inhabiting the intestinal tract.
Recent publications
Hall M, Hasegawa Y, Yoshimura F, Persson K: Structural and functional characterization of shaft, anchor, and tip proteins of the Mfa1 fimbria from the periodontal pathogen Porphyromonas gingivalis. Sci Rep. 29;8(1):1793. 2018.
Lee JY, Miller DP, Wu L, Casella CR, Hasegawa Y, Lamont RJ: Maturation of the Mfa1 fimbriae in the oral pathogen Porpshyromonas gingivalis. Front Cell Infect Microbiol.9;8:137. 2018.
Nagano K, Hasegawa Y, Iijima Y, Kikuchi T, Mitani A: Distribution of Porphyromonas gingivalis fimA and mfa1 fimbrial genotypes in subgingival plaques PeerJ 6:e5581, 2018.
Hasegawa Y, Iijima Y, Persson K, Nagano K, Yoshida Y, Lamont RJ, Kikuchi T, Mitani A, Yoshimura F. Role of Mfa5 in expression of Mfa1 fimbriae in Porphyromonas gingivalis. J Dent Res, 95(11): 1291-1297, 2016.
Funding
JSPS KAKENHI Grant Numbers 20890248, 22791783, 25861752 and 16K11466
Research grant from the Center for Advanced Oral Science, Aichi Gakuin University
Furukawa funding, Aichi Gakuin University
Yoshiaki Hasegawa is a Professor at Department of Microbiology, College of Dentistry, Aichi Gakuin University. After completing a PhD at the Department of Periodontology, College of Dentistry, Aichi Gakuin University, he pursued a post-doctoral experience at the Department of Oral Biology at University of Florida College of Dentistry (Dr Lamont’s Lab). He has been working at Aichi Gakuin University (Dr Yoshimura’s Lab) and Asahi University (Dr Murakami’s Lab) for the last 12 years to determine structure and function of P. gingivalis fimbriae.
*Please note: this is a commercial profile.