The Ebenstein lab for NanoBioPhotonics in Tel Aviv University, Israel, develops new technologies for studying genomes, an aspect of interdisciplinary research that Prof Yuval highlights here
The DNA sequence is identical in almost all cells in our body, however, each cell or tissue has a unique gene expression profile and consequently different function. These differences are a result of epigenetic mechanisms. Epigenetic marks are small chemical modifications attached to the DNA or to proteins that wrap the DNA. These modifications determine which genes will be expressed and which will be shut down. The global levels and genomic patterns of epigenetic marks are altered in many types of diseases, including cancer. These changes can be monitored and may potentially be used as biomarkers for detection and diagnosis of diseases.
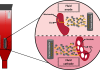
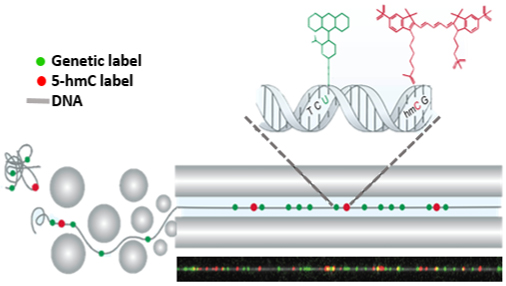
In our lab, we develop methods for labelling epigenetic marks by tagging them with a fluorescent molecule. We also label a specific genomic sequence with a different colour to generate a barcode, which helps us track the genomic location that each molecule originated from. The molecules are then stretched by squeezing them into nanochannels and imaged by a unique microscope. This process results in long DNA fibres, decorated with two colours, which can be aligned to the genome and create a map of the epigenetic marks (see figure). Since we are looking at single DNA molecules, each molecule originated from a single cell, we can study variations between cells and discover rare populations that are masked by most methods.
Hila Sharim, PhD student
In my study, I focus on the epigenetic mark 5-hydroxymethylcytosine, or 5-hmC, which was discovered in mammals less than ten years ago and its roles are not fully resolved. My goal is to use the optical detection methods developed in our lab to break up mixtures of cells into the subpopulations comprising them. Specifically, by fluorescently tagging 5-hmC, which is associated with gene expression levels in each cell, we can build a model to differentiate the specific epigenetic patterns, consequently identify each cell type.
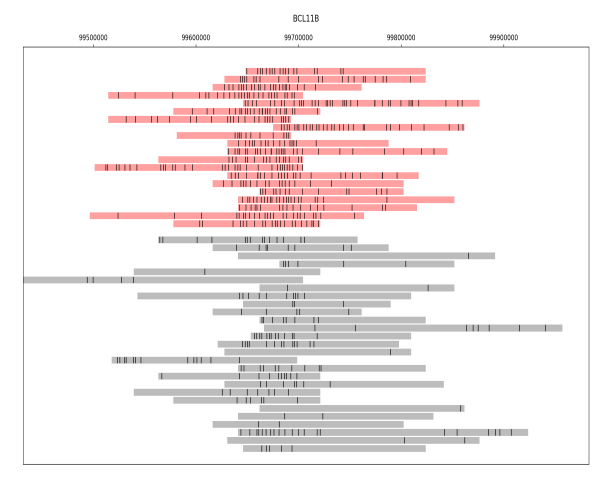
The figure presents a collection of single DNA molecules from a mixture of cells, all originating from the same genomic location. The black dots represent the detected epigenetic marks on each molecule. The molecules in the figure have been divided into two groups based on the density of their epigenetic pattern, one group shown in red and the other in grey. This division corresponds to the presence of two distinct subpopulations of cells in the mixture. The identification of unique patterns in a mixed population may be especially important for cancer diagnosis, where detecting a small subset of cells displaying irregular patterns may assist in early detection of the disease.
Since the human genome is composed of more than three billion bases, mapping the entire genome can be very expensive, especially if we want to focus our analysis in a specific region of interest. Therefore, we have developed a method for targeted enrichment of long DNA fragments, called CATCH, which utilises guided CRISPR/Cas9. Using this method, we can isolate genomic targets of up to 1-2 million bases, and still keep the DNA molecules intact with its epigenetic marks. Using this technique, we were able to isolate the breast cancer associated- BRCA1 gene with more than 200 -fold enrichment, and specifically analyse this fragment, without wasting money and resources on undesired genomic regions.
Sapir Margalit, PhD student
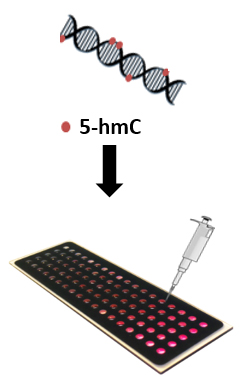
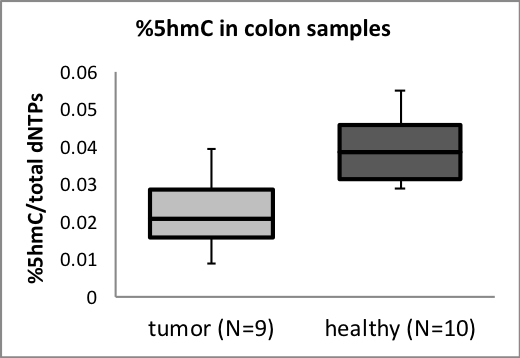
In my study, the aim is to harness epigenetic marks to develop new, easy and accessible tools for diagnosing diseases, with an emphasis on cancer. The global level of the epigenetic DNA modification 5-hydroxymethylcytosine (5-hmC) is known to significantly decrease in many cancers, and therefore, its measurement can be utilised for detection, and potentially even for prediction, of these cancers. The assay that we have developed involves sensitive fluorescent tagging and optical visualisation of this modification and can accommodate many different samples at once, providing a fast and robust analysis.
Our results indicate that this assay allows distinguishing between a healthy and cancerous colon and brain tissues based on their 5-hmC content and complement the current diagnosis workflow. Other appealing applications are detecting low 5-hmC levels present in blood, potentially enabling cancer detection using a simple blood test, and monitoring patients’ response to a given treatment at frequent intervals for personalised therapeutic decisions.
Please note: this is a commercial profile
Prof Yuval Ebenstein
Principal Investigator
Department of Chemical Physics
School of Chemistry, Tel Aviv University
Tel: +972 3640 8901
uv@post.tau.ac.il
www.nanobiophotonix.com