Frederique Lisacek from SIB Swiss Institute of Bioinformatics, discusses the experimental approaches towards Glycoscience and emphasises the need for collecting and integrating glyco-related information
Omics technology and bioinformatics
The Wikipedia definition of bioinformatics refers to the creation and advancement of databases, algorithms, computational and statistical techniques, and theory to solve formal and practical problems arising from the management and analysis of biological data. Over the past few decades, bioinformatics has played an increasingly important role in the development and expansion of -omics(1) domains that have propelled molecular biology towards the more holistic vision of systems biology. The systems approach relies on automation thereby enabling large-scale studies achieving the “-ome” (exhaustive set) goal. It also entails sophisticated modelling not in the scope of this article.
Advances in the various-omics fields have demonstrated beyond doubt the worth of well-curated and extensive data collections associated with appropriate software tools, not only to improve the analytical methods but also to support data interpretation through comparative approaches. But this progress is unevenly spread and largely depends on the level of automation. Among others, glycomics dedicated to the systematic study of glycans (also called carbohydrate or simply sugar) has lagged behind because, until recently, the development of methods for characterising the branching structures of complex carbohydrates has been slow.
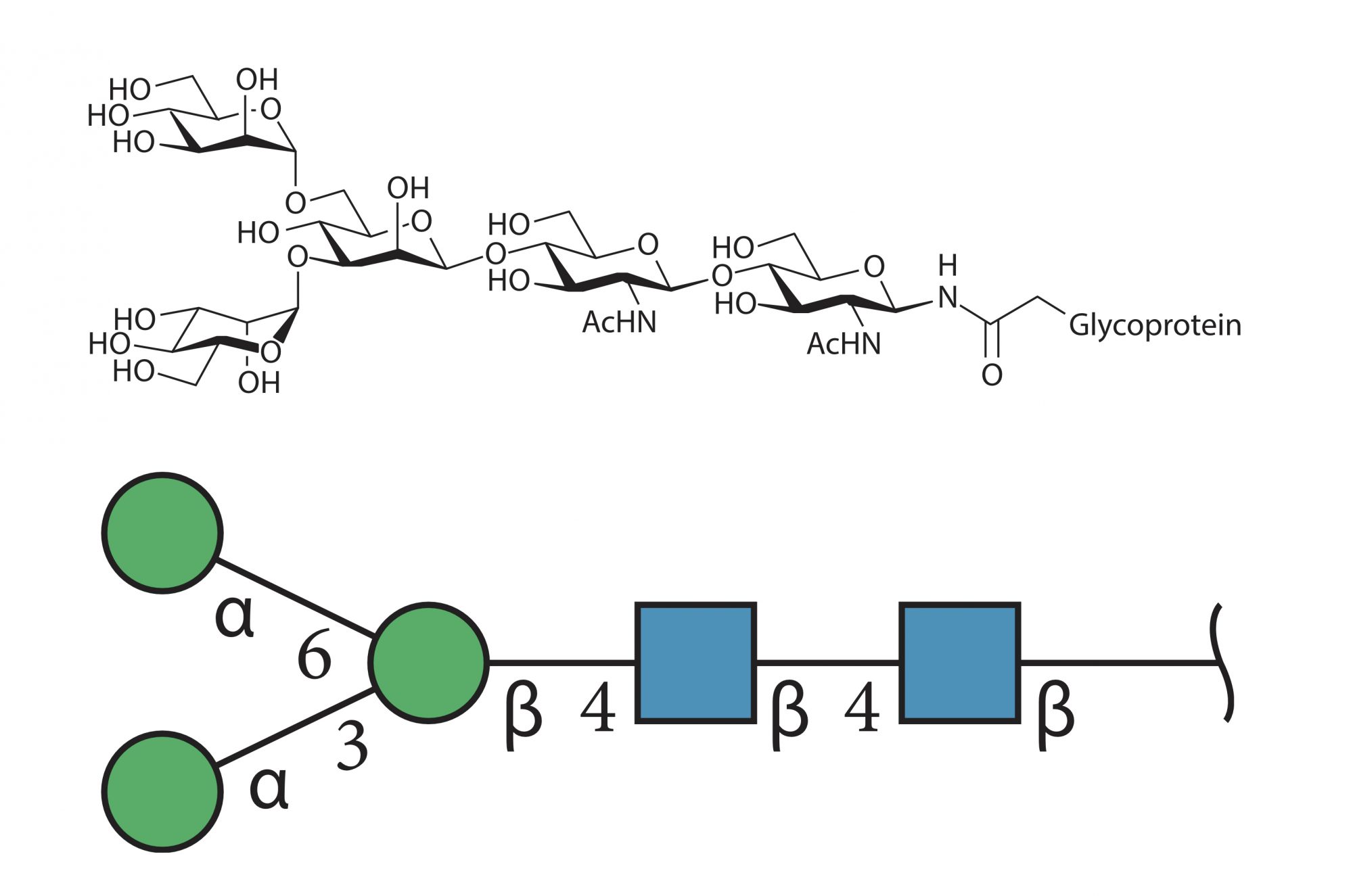
In an article of the previous issue, the notion of glycosylation of a protein was introduced. It refers to a protein on which a number of carbohydrate molecules are attached, thereby modifying or modulating its function. Full and detailed characterisation of glycan structures presented by a protein is a challenging analytical process that requires sensitive, quantitative and robust technologies to determine monosaccharide composition, linkages and branching sequences as shown below in two commonly used formats.
DNA sequencing has become a routine experiment in the last twenty years, but glycan sequencing is in its infancy, as emphasised in several published roadmaps issued by the American National Research Council in 2012, by the GlycoForum of the European Science Foundation (ESF) in 2015 and the recent also European Glyco 2030. These references unanimously point at the need for organised access to glycan-related data with the aim of supporting larger scale studies and higher throughput technology.
Heterogenous data and glycoinformatics
Glycoscience is in essence multidis-ciplinary and lies at the crossroad of chemistry and biology, impacting research in immunology, microbiology, virology, plant and marine science, to mention only the most prominent. This diversity entails an equally wide range of experimental techniques that are used in order to structurally and functionally characterise glycoconjugates (e.g., glycoproteins and glycolipids). Yet, many biologically significant events can be attributed to glycan recognition since glycoproteins, glycolipids and glycan-binding proteins are frequently located on the cell primary interface with the external environment. In other words, the interface between cells is envisaged as an intricate network of protein-protein interactions many of which are glycan-mediated.
A common approach for studying such an entanglement of molecular interactions is to attempt to separate the individual contributors to such a complex set of interactions. To do so, structural biologists tend to master X-ray crystallography, nuclear magnetic resonance (NMR) or more recently Cryo-EM (Nobel prize in 2017) techniques, while analytical chemists often rely on liquid chromatography and mass spectrometry. These “orthogonal” methodologies depend on different and usually large high-precision instruments each one requiring specific know-how. Then, functional studies capturing interactions are based on screening technology involving lighter devices such as micro-chips, which in turn necessitate another set of skills.
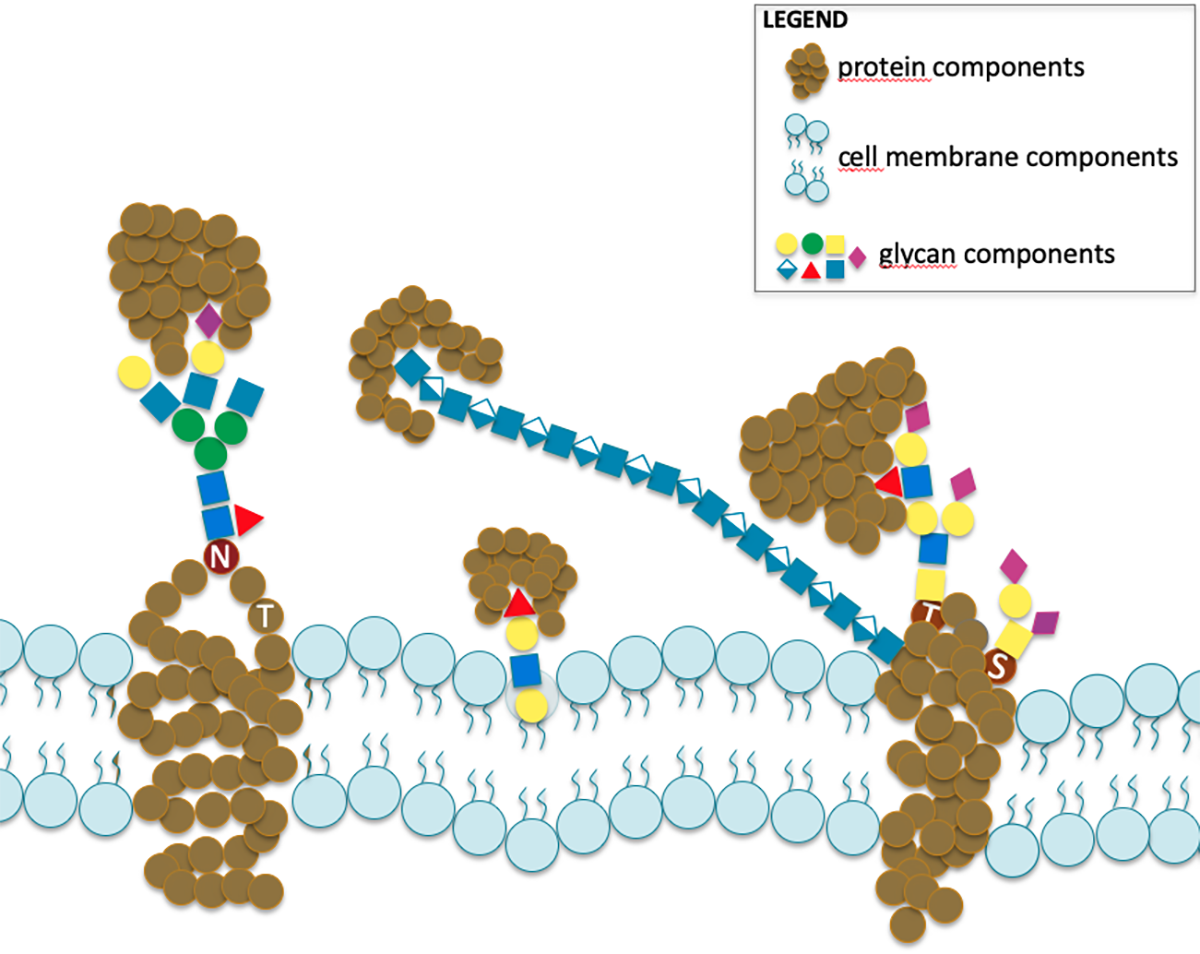
This particularly extended range of experimental approaches generate data with minimal overlap and in a diversity of often incompatible formats. Yet these data are meant to characterise the same phenomenon depicted in the image featuring cell membrane glycoconjugates (glycoproteins and glycolipids) with attached glycans bound by carbohydrate-binding proteins.
The situation as depicted so far, emphasises the need for collecting and integrating data thereby recording glyco-related information to fully characterise cell-cell communication. We undertook this task and provide a range of glyco-related databases and software tools available in the “glycomics” section of the portal of the SIB Swiss Institute of Bioinformatics called Expasy.
Cooperative science
The Alliance Campus Rhodanien (ACR) is a network created in 2017 to support synergies between six academic institutions(2) naturally brought together by geographical (Alps and Rhone region) and linguistic proximity. ACR aims at expanding joint cross-border scientific research and innovation as well as amplifying the participation of ACR members in European cooperation programmes.
In a 2018 competitive bid, a glycoinformatics project led by three teams of Geneva, Grenoble and Lyon universities was selected. The initial goal was to interconnect the three partners’ respective databases which represent different though complementary and related fields in glycobiology: 2D glycan structures and glycoproteins/ Geneva (mainly originating from mass spectrometry data), 3D glycan structures and glycan-binding/Grenoble (mainly originating from X-ray crystallography data), and extracellular matrix protein -carbohydrate complexes/Lyon (mainly originating from micro-chip data). This integration required the use of standards promoted in glycoinformatics and bioinformatics according to the FAIR (Findable, Accessible, Inter-operable, and Re-usable) principles promoted in Open Science. Such an approach was still rare in glycoscience a few years ago, but fortunately, is now spreading in glycoinformatics.
In a second phase, the project was shaped toward the construction of functional 3D networks of protein-glycan interactions, specific to cells, tissues or pathologies, which requires the integration of structural proteomic and glycomic data (Grenoble/Lyon/Geneva). The federating activity of the Geneva team via the Expasy glycoinformatics portal was key to the ACR project, which was aligned with the objectives of centralisation and unification that guide the development of the portal. Further, the common involvement of two of the three teams in the Carbohydrate Metrology Action (EU/FET-OPEN CarboMet project), which brings together a number of European academic groups and companies with a networking goal, was another asset.
This multi-level cooperative approach illustrates the rising trend in glycoscience of reaching out to other omics following recent progress in carbohydrate structure resolution and synthesis as well as functional screening methods. The input of glycoscience is gradually recognised as the roles of glycans and glycoconjugates are manifold and revealed in various medical, biochemical and biotechnological applications.
To complement the virology illustration of the previous issue article, at another level, it should be noted that the bacterial balance in the gut microbiome crucially involves carbohydrates. A tight knit of interactions between host glycoconjugates located on the gut mucus (coated with glycans), bacterial catabolic enzymes and bacterial surface proteins, needs to be disentangled to understand the detrimental effects of a growing number of intestinal diseases. For instance, it was shown that a low-fibre diet (therefore low in carbohydrate) promotes the expansion and activity of mucus-degrading bacteria which feed on the gut glycan protective coating.
In the end, our collaborative strategy is geared toward providing high quality, reliable, well-referenced, and accurate data to the Life Science community.
- The -ome suffix refers to a collection that is complete such as, all genes make a genome, all proteins make a proteome, etc.
- The Universities of Geneva (UNIGE), Lausanne (UNIL), Grenoble- Alpes (UGA) and Lyon (UdL), as well as the University of Applied Sciences Western Switzerland (HES-SO). The University of Savoie Mont Blanc (USMB) joined the network in 2020.
Please note: This is a commercial profile