Richie Kohman, Synthetic Biology Platform Lead, Wyss Institute at Harvard, tells us all about Next Generation Sequencing in analysing synthetic biology signatures in brain tissue
Next Generation Sequencing (NGS) is one of the most advanced methods in biotechnology, having experienced a million-fold cost reduction over the past two decades. Once a technique only applicable to samples prepared for analysis on commercial sequencers, NGS is now being applied directly in biological samples to investigate the spatial locations of nucleic acids. Most commonly, the spatial expression of genes as determined through RNA localisation is performed, however, there is a recent movement to analyse bioengineered molecules in addition to natural ones.
Using the tools of Synthetic Biology, researchers are now writing molecular signatures into biological systems and using NGS to read them out. This general approach has a unique advantage. Many processes in biology do not leave easy-to-analyse markers along the way as they progress. Because biological systems are immensely complicated with seemingly endless biochemical processes occurring over time, researchers often do not have molecular markers attributed to the process they are trying to examine. Synthetic Biology offers a means to tag, or write-in, desired molecules at specific times or locations within cells, and, if properly executed, the readout of these tags can allow challenging biological phenomena to be studied more effectively.
Brain mapping
One scientific task that has been especially challenging to tackle is high-resolution brain mapping. The mammalian brain is intricately connected with neurons forming circuits locally as well as over long distances. For example, each neuron in the mammalian cortex (which has 90,000 neurons per cubic millimetre) is known to form connections with approximately 8000 other cells.
Understanding how these cells are wired would not only give insight into fundamental brain processes such as cognition or the storage of memories but could also lead to a better mechanistic understanding of disease progression and the discovery of better treatments for neurological and psychiatric disorders. Thus far electron microscopy has been the primary tool used to uncover high-resolution brain connectivity. Tracing cell paths over thousands of thin brain sections has provided accurate anatomical reconstructions however the process is laborious and challenging to scale to large volumes of brain tissue. To overcome this limitation, new research efforts are being investigated.
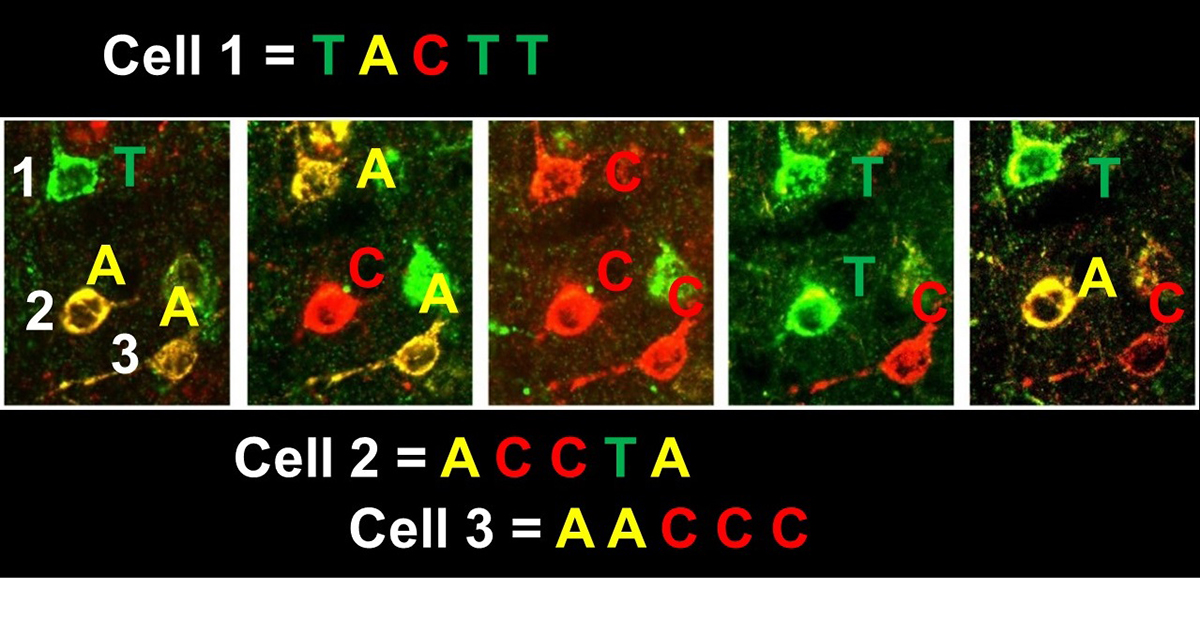
Several researchers are utilising NGS to explore brain anatomy. As one of the earliest figures in this field, Antony Zador from Cold Spring Harbor Labs outlined this strategy in a 2012 essay(1). Here it was commented that the low cost and high sequencing scale of NGS make it an appealing technique to apply to new scientific challenges, such as mapping brain circuits. If unique nucleic acid sequences could be assigned to individual neurons and these sequences could be positioned at points of cell connectivity, then NGS readout of these “barcodes” could discover connectivity at a much higher rate than previous electron microscopy-based methods.
The invention of the MAPseq in 2016 applied this concept to studying neuron projections in the mouse(2). In this approach, RNA barcodes were expressed throughout neurons in the brain through a viral injection. After infection, the brain was divided into small volumes that were each processed for NGS. Because the identity of the barcodes could be mapped to the locations of tissue they were taken from, MAPseq was able to analyse thousands of single neuron projects in approximately one week’s time.
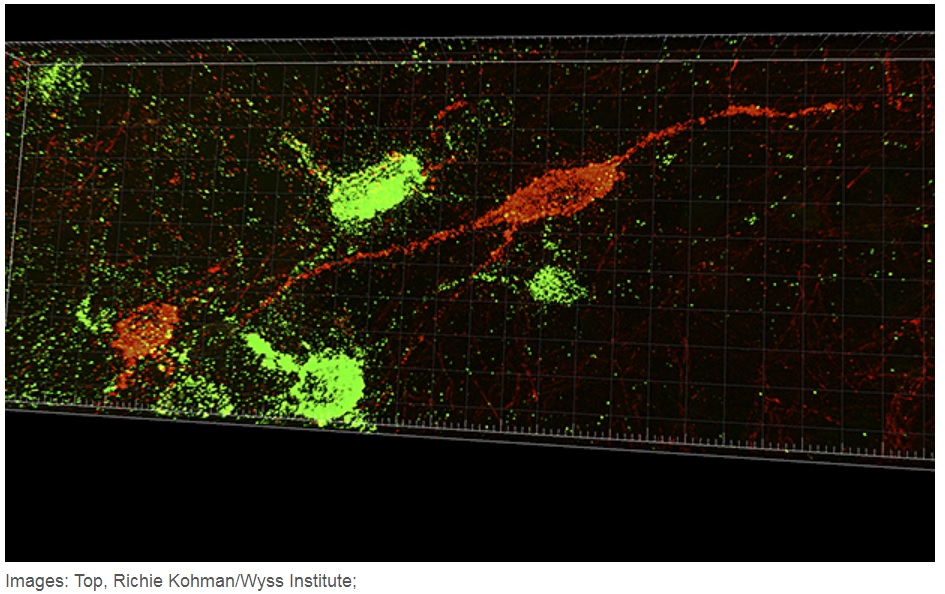
As interest in brain mapping has grown, so has the number of research initiatives dedicated to this field. Recently the IARPA MICrONS (Machine Intelligence from Cortical Networks) project was created to revolutionise machine learning by reverse-engineering the algorithms of the brain. One MICrONS team lead by Professor George Church and Dr Richie Kohman from the Wyss Institute laboratory at Harvard Medical School aimed to use cutting-edge in situ NGS techniques(3,4,5) to analyse cellular barcodes, such as those used in MAPseq. This task necessitated the creation of new in situ variations such as BARseq(6) and eventually led to the invention of super-resolution barcode sequencing as first shown by Kohman in 2017 (Figure 1).
This advent of high-resolution mapping of cellular connectivity has the potential to propel the field forward in scale. With further advances, it will one day be possible to obtain connectivity diagrams of multiple whole organs and enable comparative connectomics.
By being able to compare tissue of healthy brains with diseased ones, it will be possible to uncover connectivity differences specific to the disease and potentially identify new therapeutic options. Taken together, the readout of synthetic biology signatures in the brain has the potential to revolutionise the analysis of brain architecture. This example demonstrates the power NGS as a tool that can be applied to new research areas not initially imagined for the technology and also highlights the creativity of the scientists and engineers who venture into these new areas.
References
1) Zador A.M. et al. Sequencing the Connectome. PLOS Biology 2012, 10(10): e1001411.
2) Kebschull, J. M. et al. High-Throughput Mapping of Single-Neuron Projections by Sequencing of Barcoded RNA. Neuron 2016, 91 (5), 975-987.
3) Turczyk, B. M. et al. Spatial Sequencing: A Perspective. J Biomol Tech 2020, 31 (2), 44-46.
4) Lee, J. H. et al. Highly multiplexed subcellular RNA sequencing in situ. Science 2014, 343 (6177), 1360-1363.
5) Lee, J. H. et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc 2015, 10 (3), 442-458.
6) Chen, X. e t al. Efficient in situ barcode sequencing using padlock probe-based BaristaSeq. Nucleic Acids Res 2018, 46 (4), e22.
7) https://spectrum.ieee.org/biomedical/imaging/ai-designers-find-inspiration-in-rat-brains
Please note: This is a commercial profile
© 2019. This work is licensed under CC-BY-NC-ND.