Dr. James E Goldman and Dr. Osama Al-Dalahmah from the Department of Pathology and Cell Biology at Columbia University, provide an in-depth perspective on Huntington’s disease (HD) research
Huntington’s disease (HD) is a progressive, inherited, neurodegenerative disorder, characterised by abnormal, involuntary movements and varying degrees of cognitive difficulties, such as memory problems or difficulties making decisions or organising their lives, and emotional problems, such as anxiety and depression. HD is caused by mutations in the Huntington gene (HTT), which produces a mutant protein. The disease is genetically dominant; that is, only one of our two HTT genes needs to be mutated to produce HD, and each child of a person with HD has a 50% chance of receiving the mutant gene.
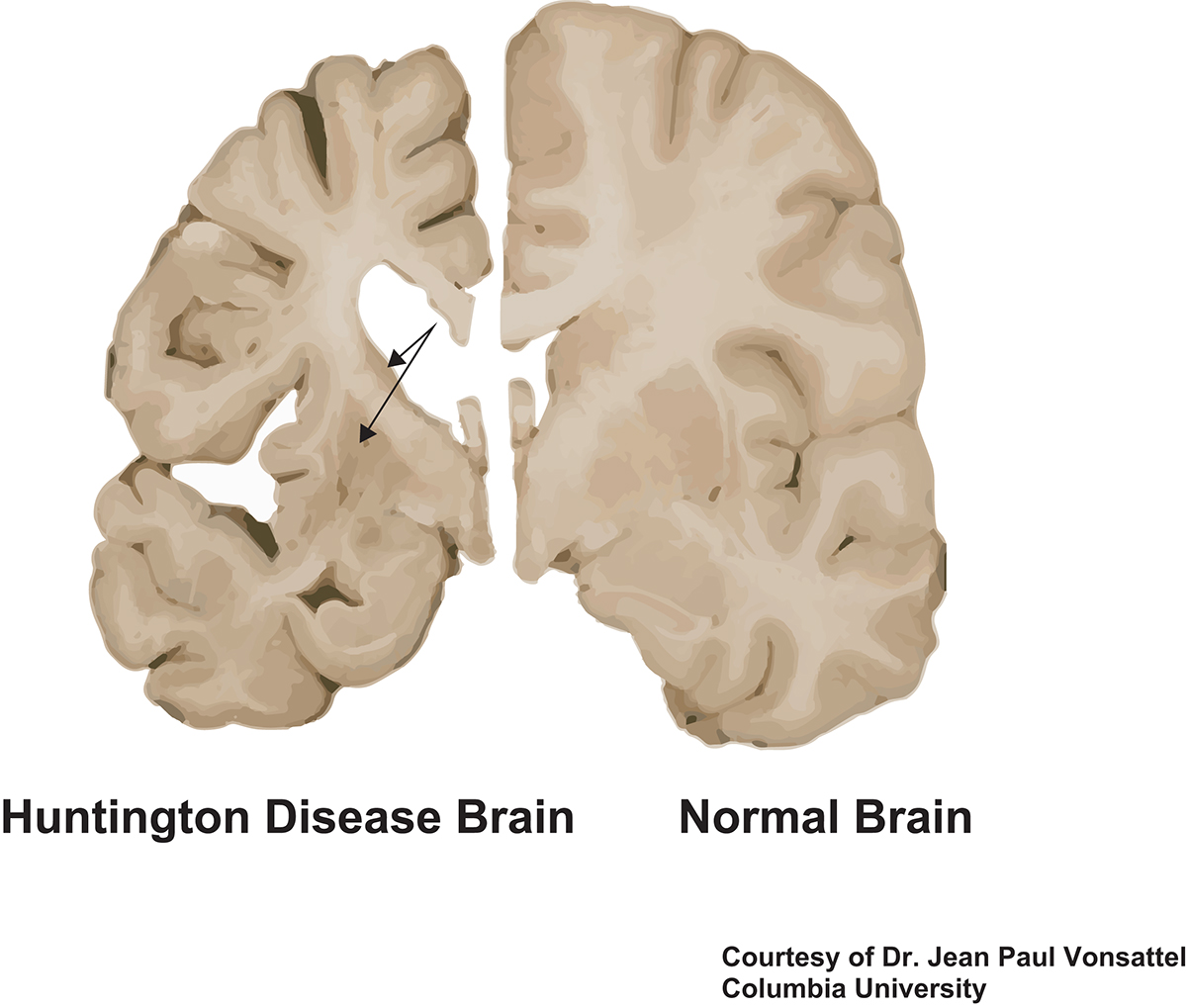
HD brains have been examined for decades by neuropathologists. There is severe loss of neurons in the striatum, an area in the central part of the brain that plays a large role in regulating smooth movements. Areas of the cerebral cortex also show neuronal loss. There is loss of myelin, the fatty sheath that surrounds and insulates axons, the neuronal processes through which neurons communicate with each other. Other cells in the brain, including astrocytes, glial cells that support neuronal function and health, and microglia, the immune cells of the brain, are also affected.
Molecular mechanisms causing HD
Although neuropathology has given us essential clues to the structural changes in HD brains, it has not provided us with an understanding of the molecular mechanisms causing the disease. In recent years, investigators have been able to produce the HTT mutation in mice and cells grown in tissue cultures. These studies have begun to reveal important mechanisms by which the mutation produces this remarkable pathology. Also, new technologies have emerged to allow us to investigate molecular changes in the human brain. One of these is single nucleus RNA sequencing (snRNAseq).
With this approach, we isolate cell nuclei from different areas of HD brains, using fresh frozen brain tissue taken at autopsy and stored at a very low temperature in the New York Brain Bank at Columbia University Medical Center. Cell nuclei contain the cell’s DNA and also the RNA made from the genes encoded in DNA. RNA is then transported from the nucleus into the cell cytoplasm, where it is “read” and translated into cell proteins. Note that not all genes are expressed in all cell types, so, for example, the expression of genes in liver cells are different from those expressed in muscle cells.
Furthermore, the genes expressed in normal cells must be significantly different from those expressed by the same cells in a disease. The snRNAseq technology allows us to capture RNAs in nuclei and then to determine exactly which RNAs are present in single nuclei. Thus, we obtain a list of genes expressed in all of the different cell types and compare those in HD brains to those in the brains of people without neurological disease. We can even compare these gene expression profiles to the same cell types in other neurological diseases, such as Alzheimer’s and Parkinson’s diseases. Note that in mouse models of disease one can isolate cells, not just nuclei, from freshly removed brains, but that is impossible in frozen tissues. It is possible to do this to some extent from a fresh human brain, but more difficult, and one cannot use already banked material.
Another feature of snRNAseq is that we can examine different regions of the brain. For example, in HD and the area of the central part of the brain, the striatum, is more affected than other areas such as the cerebral cortex. That is, earlier in the disease when there is a much great loss of neurons in the striatum than in other areas. We can now compare gene expression profiles of different cell types in different areas, discriminating these differences.
How can single-cell studies help us understand more about HD?
In the brain of patients with neurodegenerative diseases, not all cells show pathological changes simultaneously. These diseases progress over years and sometimes decades. Thus, in looking at single cells, we find a mixture of those that are severely changed and those that are much more moderately changed. The cells along this spectrum have different gene expression patterns, and by examining these patterns we can infer some of the earliest changes in the disease. This is important because therapies would be most effective if offered early in the course of a progressive disease, even before signs and symptoms begin. As one example of this, we have found that astrocytes in the HD brain can be grouped by their gene expression patterns into those we believe are farther along in a pathological spectrum than others.
Furthermore, gene expression changes in astrocytes in the striatum are different from those in the cortex of HD brains. We find that early responses of astrocytes involve the increase in the expression of genes that provide protection from heavy metals, such as iron and copper, heavy metals that are increased in the HD brains, whereas later stages of the astrocyte pathology has lost these protective genes. Neuropathological studies point out that even within one area, such as the striatum or cortex, there is heterogeneity in pathology. Thus, pathology in the striatum proceeds in a dorsal (upper) to ventral (lower) gradient. Examining cells from different striatal regions may also give us insights into changes in gene expression as HD progresses.
Modern technologies add more to snRNAseq. We can now pick a series of RNAs that are increased or decreased in the HD brain and apply probes for those RNAs to tissue sections of the brain to ask if the cells with the altered gene expressions are distributed randomly over the tissue or whether certain areas of the striatum or cortex are affected. We are certain that there is not a random distribution. Again, this visualisation can give us insights into where the earliest changes are in the brains.
The discovery of changes in gene expression in HD and other diseases is giving the scientific community new and important insights into the molecular mechanisms that underlie these disorders. Particularly, finding early changes in gene expression may well lead us to better therapies.
*Please note: this is a commercial profile