Didier Andrivon from INRA details why an intimate knowledge and understanding of the potato late blight pathogen and its evolution are key towards sustainable control
Phytophthora infestans, the causal pathogen of potato late blight, is known to be a versatile enemy. Its ability to constantly generate new variants is certainly one of the main assets of this highly epidemic parasite for causing recurring threats on potato and tomato crops worldwide.
The recent EuroBlight workshop, held in York (UK) from 12th to 15th May 2019, once again highlighted the new emergences of clonal lineages of P. infestans going on in Europe, and reflected on their immediate and longer-term consequences for sustainable management of the disease. It also shed further light on global migrations of European lineages and on the issues these pose for blight control globally.
Ongoing changes in European P. infestans populations…
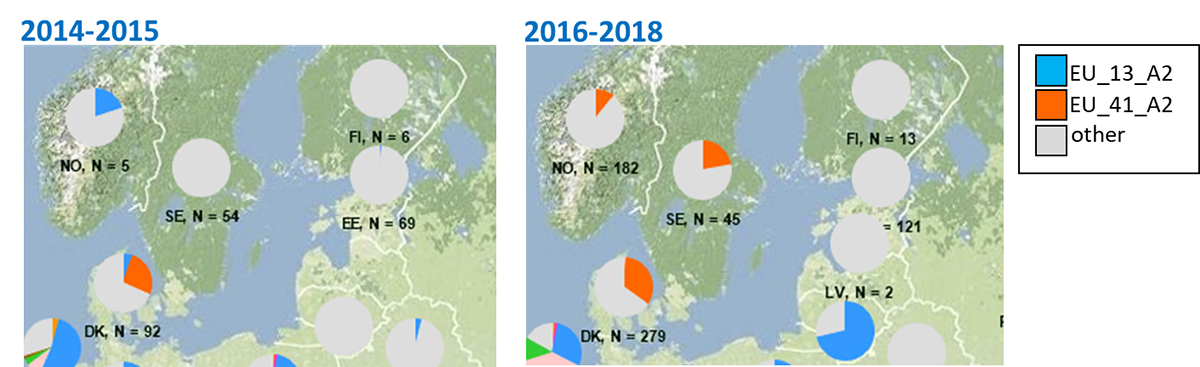
As yet rare genotypes EU_36_A2 and EU_37_A2, first spotted in the northern part of the Netherlands in 2013/2014, started spreading over north-western Europe from 2016, and replaced earlier lineages, such as EU_13_A2 and EU_6_A1 there. Meanwhile, genotype EU_41_A2 established itself in sexual, Nordic populations over the past three years (Figure).
The precise reasons for such rapid population invasions are yet to be fully elucidated, but some phenotypic characteristics of the newcomers are probably part of the equation. Indeed, recent reports confirmed that genotype EU_37_A2 is insensitive to fluazinam, a major late blight fungicide. Investigations made within the frame of the European project IPMBlight2.0 also indicated that genotype EU_36_A2 was among the most aggressive of current populations and that genotype EU_41_A2 had the ability to overcome many race-specific resistance genes available to breeders or present in current potato cultivars.
Despite these advances and new, quantitative data on major phenotypic traits and their distributions across genotypes and geographical areas, our scientific community still lacks some critical information to turn population information into an asset for predictive management of blight outbreaks and emergencies. In particular, the elusive connection between genotypic and phenotypic variation in P. infestans and the impossibility do correctly quantify the response of pathogen populations to selection for want of accurate, geo-referenced databases on fungicide and cultivar distribution, leave many tentative explanations for the rapid changes observed in population structures untested.
…but also worldwide
The same methodologies were also successfully applied to track the recent spread of more ancient European P. infestans genotypes over their worldwide expansion. Of particular significance are the recent discoveries of the presence of genotype EU_2_A1 in Latin America, eastern Asia or eastern sub-Saharan Africa with major consequences for control efficacy.
The main question open now is ‘what makes different clonal lineages invasive in different environments’ ? The phenotypic traits measured in the lab are only part of the answer, for two reasons. First, they are measured under optimal conditions for the pathogen – which are not constantly present in the field. Therefore, pathogenicity data under sub-optimal environmental conditions would be needed for a better prediction of pathogen fitness in the field. However, acquiring such data is technically challenging and time-consuming, explaining why they are currently so scant. Second, the laboratory tests reveal extensive variation not only between lineages but also among individuals belonging to the same lineage. This intra-clone variation, therefore, allows selection of more or less pathogenic within each clonal lineage, without affecting the balance between lineages in the general population. Such intra clonal selection needs to be better documented and quantified, so it can be integrated into population dynamics models to simulate invasive dynamics more accurately, according to local competitors, environmental conditions and the resistance features of host cultivars.
Such integrative models are currently under development. Because they are based on biological traits of the pathogen rather than on local cropping practices, they will apply to all types of potato production situation – conventional or organic, in developed as in developing countries. They will certainly help us turn population data into helpful tools not only to understand ongoing changes as they are monitored (for instance, via the Euroblight population surveys) but also to anticipate them to the advantage of growers. Indeed, they should be the key to better adjust the choice of cultivars, of field applications of fungicides or of biocontrol products and the timing of these applications, depending on the local characteristics of pathogen populations present. This is the basis of IPM 2.0, a key to a more sustainable control of major diseases such as late blight.
Didier Andrivon
Research Director (Senior Scientist)
INRA
Tel: +33 (0)223 485 193
https://www6.rennes.inra.fr/igepp
Please note: This is a commercial profile











